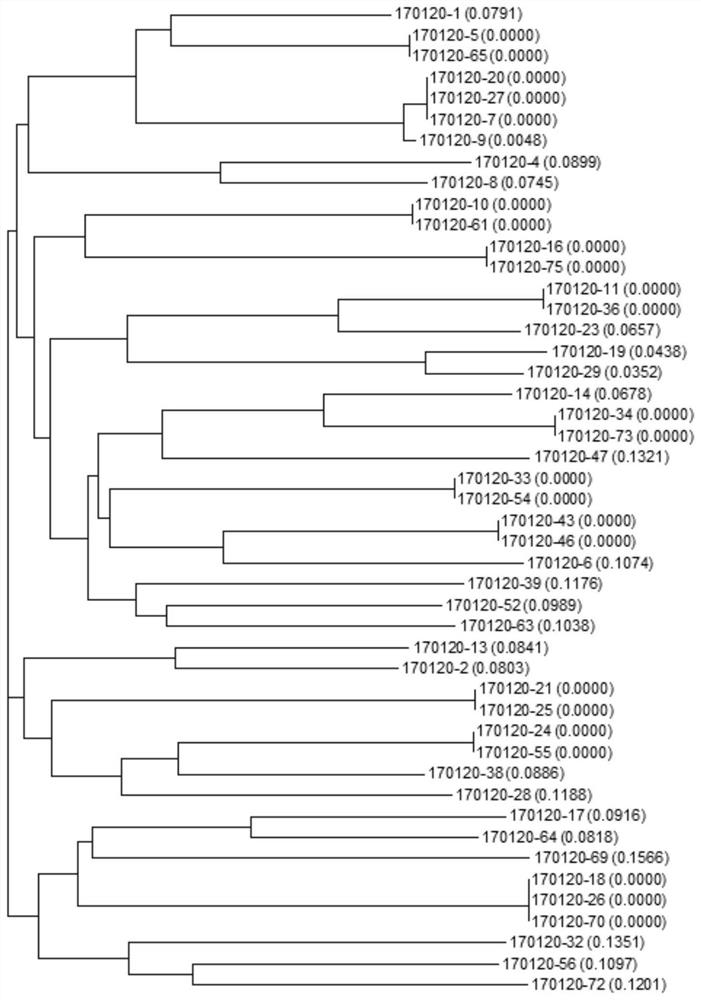
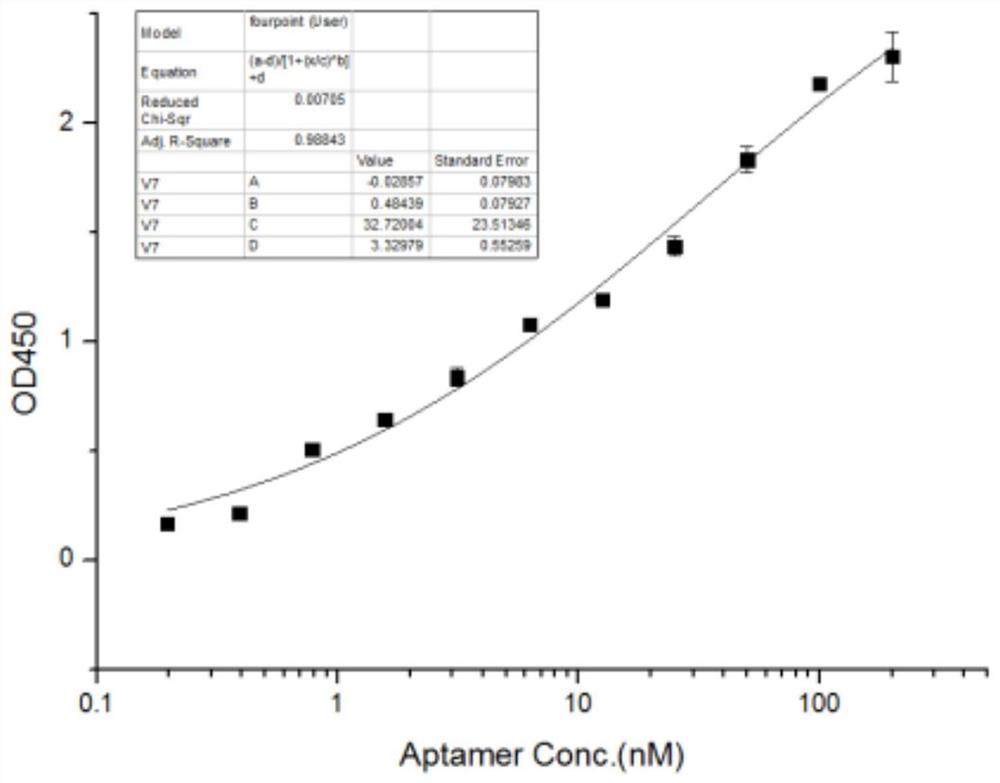
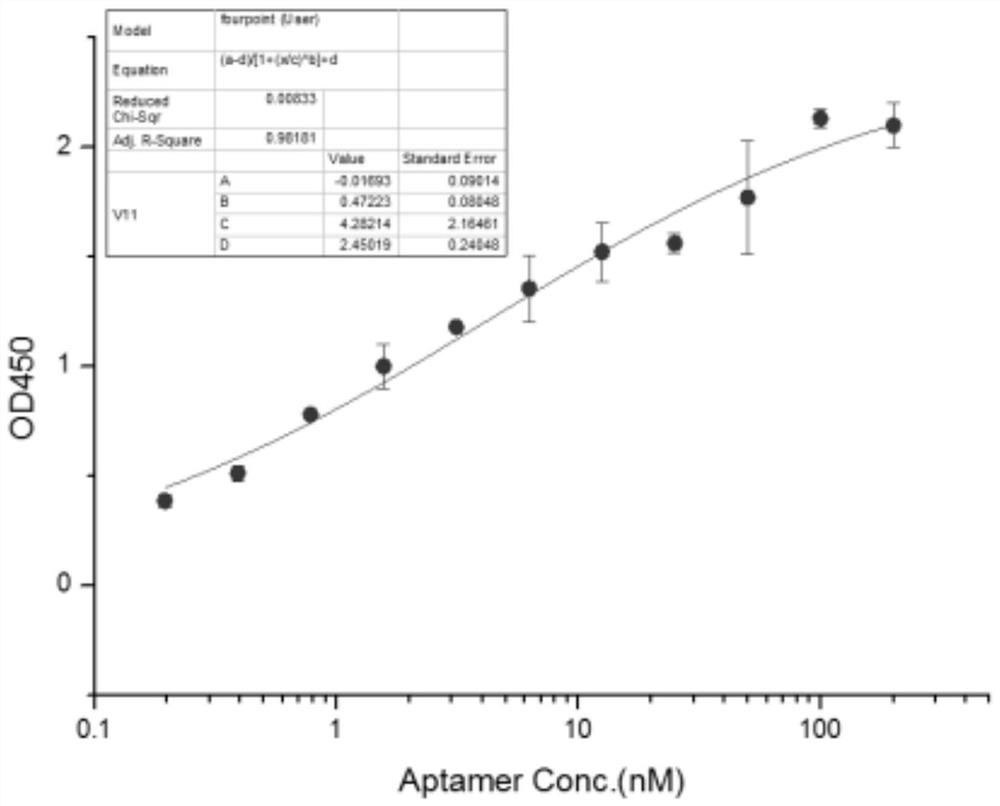
EV71 single-stranded DNA aptamer and chemiluminescent detection kit for detecting enterovirus 71 using double aptamers
A chemiluminescence reagent, EV71 technology, applied in the field of EV71 virus qualitative detection, achieves high specificity, simple and fast detection operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Screening of EV71 aptamers:
[0036] 1) Main reagents:
[0037] Binding Buffer: 1*PBS, 5mM MgCl 2 , 1% BSA, 1μg / ml tRNA, 0.02% tween-20, 5mM imidazole Washing Buffer: 1*PBS, 5mM MgCl 2 , 0.02% tween-20
[0038] Table 1 Screening library and primer sequences
[0039]
[0040] 2) The first round of SELEX
[0041] a.library was dissolved in water to a final concentration of 20 μM.
[0042] b. Configuration extension system
[0043]
[0044] The extension program is as follows: 95°C for 60s, 58°C for 60s, 72°C for 60min
[0045] c. Add the product to SA agarose bead and incubate at room temperature for 10 minutes to wash away unbound sequences.
[0046] d. Add 150mM NaOH and incubate at room temperature for 5min, aspirate the supernatant, and add 300mM HCl to neutralize.
[0047] e. Using water as a control, Nanodrop measures the concentration of modified single-stranded DNA.
[0048] f. Denaturation at 95°C for 5 minutes, add Binding Buffer to 1ml after ice ...
Embodiment 2
[0082] Aptamer affinity assay (ELISA):
[0083] a. Dilute the protein with PBS to a final concentration of 200ng / ml, set 2 replicate wells for each experimental group, add 50μl to each well. After covering the plate with sealing film, keep overnight at 4°C.
[0084] b. Washing: fill each well with Washing Buffer, let it stand for 5 seconds, pour off the washing solution, and repeat 3 times. Pat dry one last time.
[0085] c. Add 150 μl of PBS containing 1% BSA to each well to block, and incubate at room temperature for 1 hour.
[0086] d. Plate washing: the operation is the same as step b.
[0087] e. Dilute aptamer to the target concentration with Binding Buffer.
[0088] f. Take the coated strip and add 50 μl to each well. Incubate at room temperature for 1h.
[0089] g. Plate washing: the operation is the same as step b.
[0090] h. Dilute Streptavidin-HRP with Aassay Diluent 1:200. Add 50 μl to each well. Incubate at room temperature for 0.5h.
[0091] i. Washing...
Embodiment 3
[0096] Aptamer Specificity Assay (ELISA):
[0097] Referring to the experimental method in Example 2, four coating concentrations of 100ng / ml, 200ng / ml, 500ng / ml, and 1000ng / ml were set up, and the amount of aptamer fixed in each well was 200nM. The result is as Figure 5 As shown, the BSA protein at each coating concentration did not bind to the aptamer. Under the condition of higher coating concentration, the aptamer can also bind to Coxsackie virus A16 (cox) to a certain extent, but the binding ability is significantly lower than that of enterovirus 71 (EV).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com