Pre-treatment method for detecting bacterial metabolomics through gas chromatography-mass spectrometer (GC-MS)
A GC-MS, metabolomics technology, applied in microorganism-based methods, biochemical equipment and methods, treatment of microorganisms with electricity/wave energy, etc. Coverage, failed detection of metabolic compounds, etc., to maximize the number, the best vitality, and the best metabolic state
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
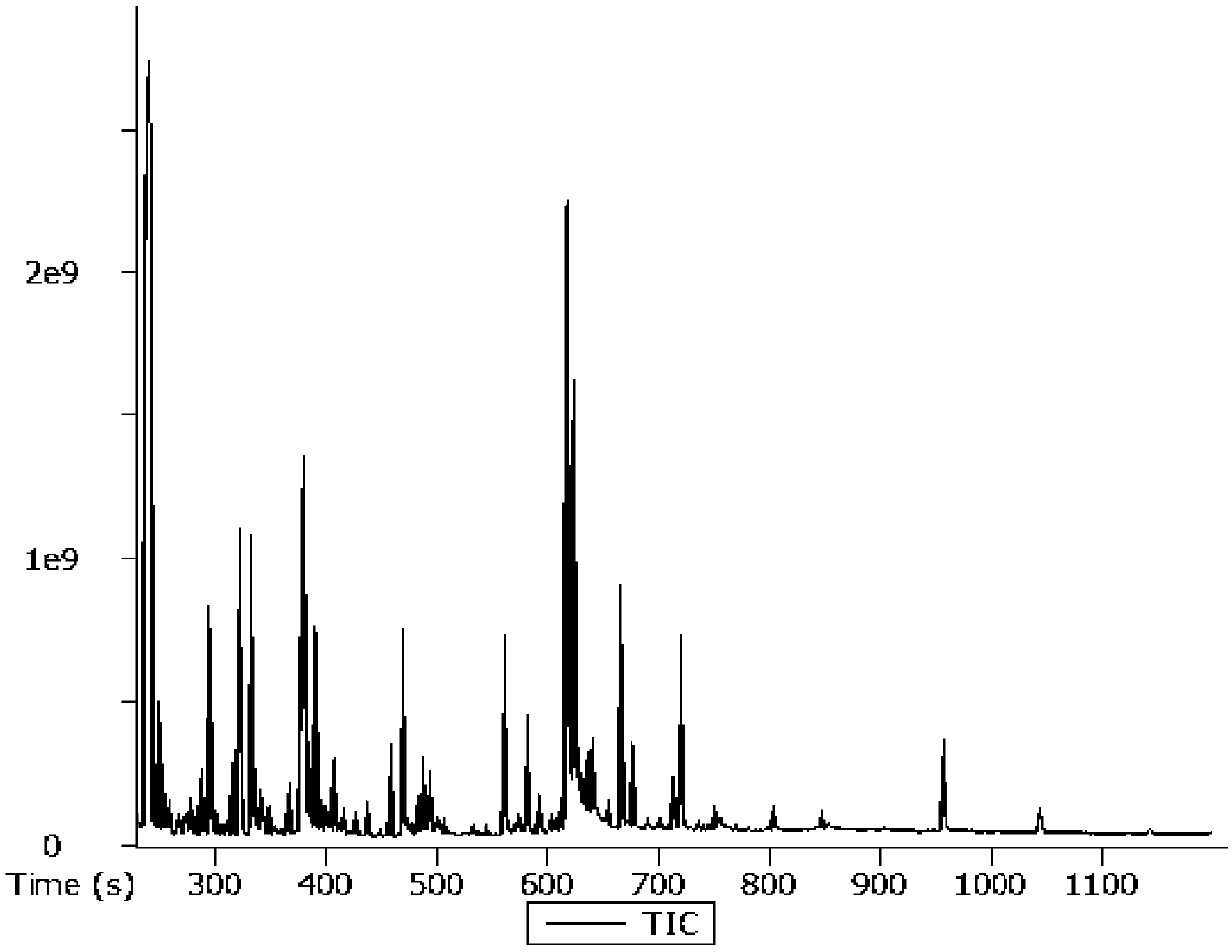
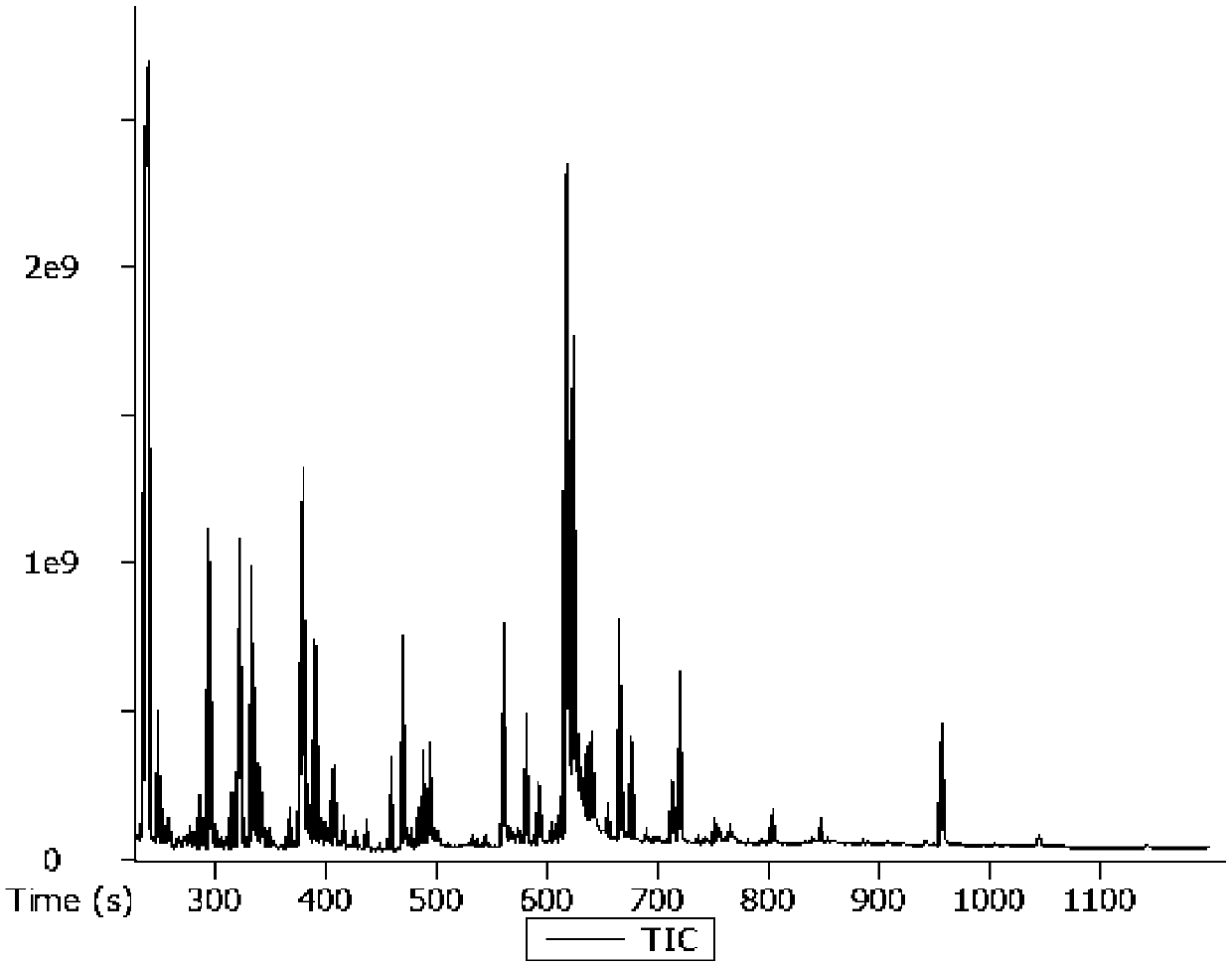
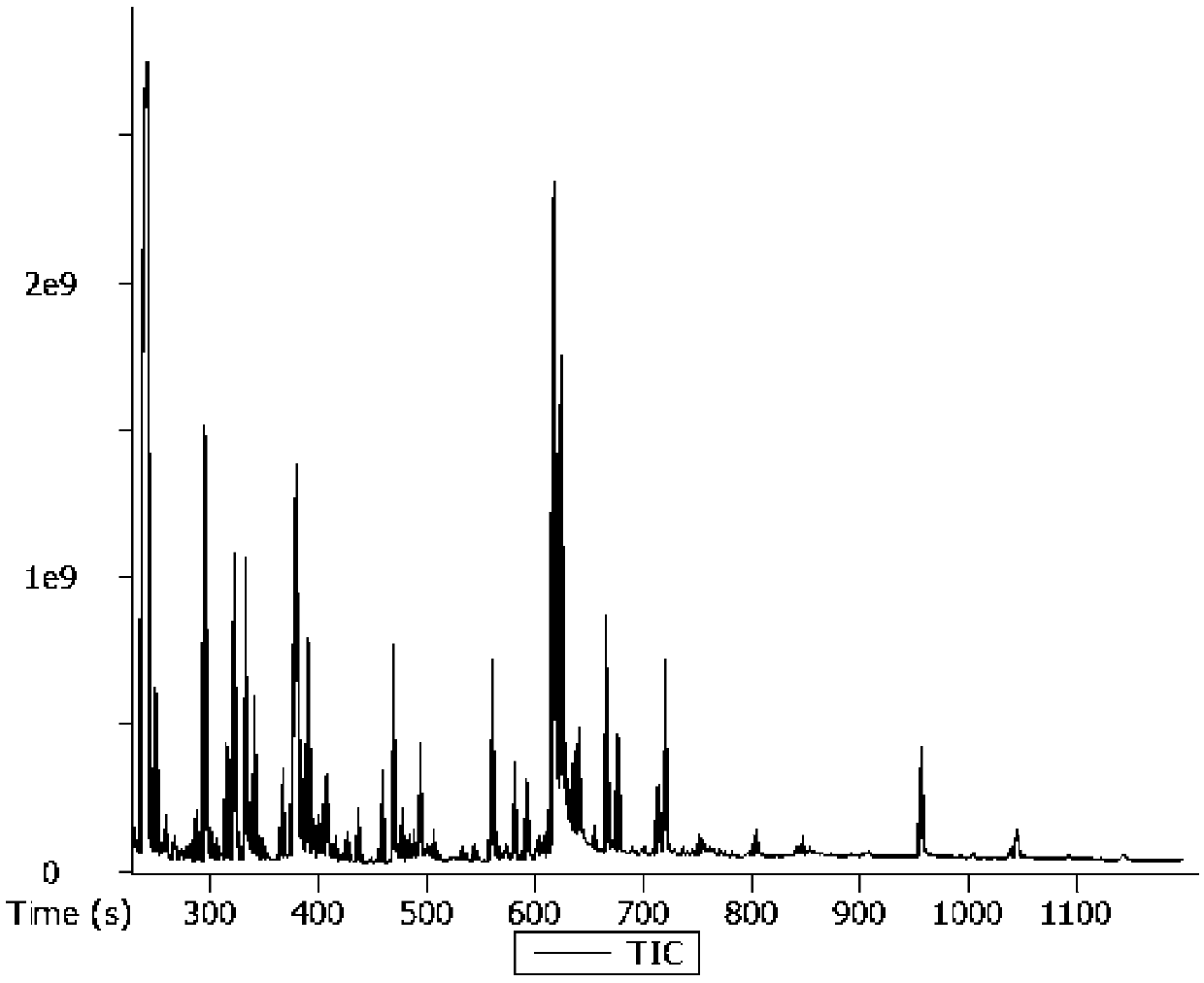
Image
Examples
Embodiment 1
[0042] Pretreatment method for Escherichia coli metabolome analysis:
[0043] (1) Escherichia coli liquid culture:
[0044] Escherichia coli was inoculated on nutrient agar medium, cultured at 37°C for 12-14 hours, stored at 4°C, picked a single colony on the plate with a sterile inoculation loop and inoculated into nutrient broth liquid medium, placed the test tube in Cultivate in a constant temperature shaker (80-120r / min) at 30-37°C for 12-16 hours to obtain an E. coli liquid sample with a McFarland turbidity of 0.4-0.6;
[0045] (2) Ice methanol-sterile water quenching solution quenches E. coli cells:
[0046] ① Put the sample of the bacterial liquid test tube and the ice quenching solution (mixed with methanol and sterile water in a volume ratio of 3:2, and pre-cooled in a -20°C refrigerator) on the ice-water mixture;
[0047] ② Add the methanol-water quenching solution to the E. coli bacterial liquid sample, store in a centrifuge tube, and the volume of the sample and ...
Embodiment 2
[0060] Salmonella metabolome analysis pretreatment method:
[0061] (1) Salmonella liquid culture
[0062] Inoculate Salmonella on nutrient agar medium at 37°C for 12-18 hours, store at 4°C, pick a single colony on the plate with a sterile inoculation loop and inoculate it into nutrient broth liquid medium, place the test tube at 30- Cultivate in a constant temperature shaker (80-120 r / min) at 37°C for 12-18 hours to obtain a Salmonella liquid sample with a McFarland turbidity of 0.4-0.6.
[0063] (2) Ice methanol-sterile water quenching solution quenches Salmonella cells:
[0064] ① Place the sample of the bacterial liquid test tube and the ice quenching solution (mixed with methanol and sterile water in a volume ratio of 2:1, and pre-cooled in a -20°C refrigerator) on the ice-water mixture;
[0065] ② Add the methanol-water quenching solution to the Salmonella liquid sample and store in a centrifuge tube. The volume of the sample and the quenching solution is 1:2;
[0066...
Embodiment 3
[0077] Pretreatment method for metabolome analysis of Staphylococcus aureus
[0078] (1) Liquid culture of Staphylococcus aureus
[0079] Staphylococcus aureus was inoculated on nutrient agar medium, cultured at 37°C for 12-16 hours, picked a single colony on the plate with a sterile inoculation loop and inoculated into nutrient broth liquid medium, and placed the test tube at 30- Cultivate in a constant temperature shaker (80-120 r / min) at 37°C for 12-16 hours to obtain a Staphylococcus aureus liquid sample with a McFarland turbidity of 0.4-0.6.
[0080] (2) ice methanol-sterile water quenching liquid quenches Staphylococcus aureus cells:
[0081] ① Put the sample of the bacterial liquid test tube and the ice quenching solution (mixed with methanol and sterile water in a volume ratio of 3:1, and pre-cooled in a -20°C refrigerator) on the ice-water mixture;
[0082] ② Add the methanol-water quenching solution to the Staphylococcus aureus liquid sample and store in a centrifu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com