Mutator strain with loss of correcting function of escherichia coli dnaQ gene, and preparation method and applications thereof
A technology of Escherichia coli and gene correction, applied in the field of biotechnology engineering, to reduce working time cost and improve efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
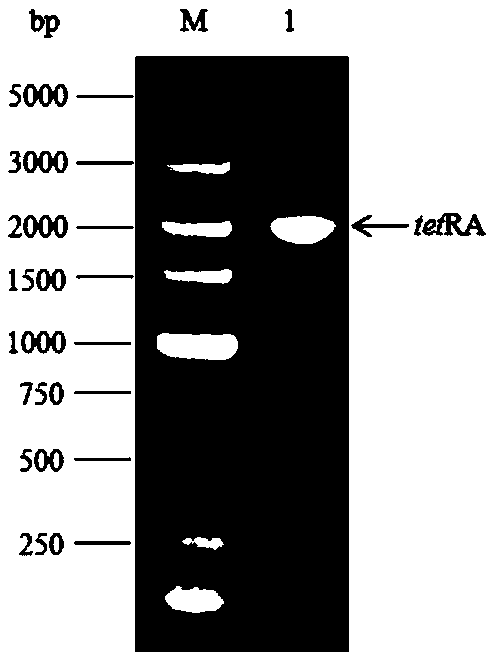
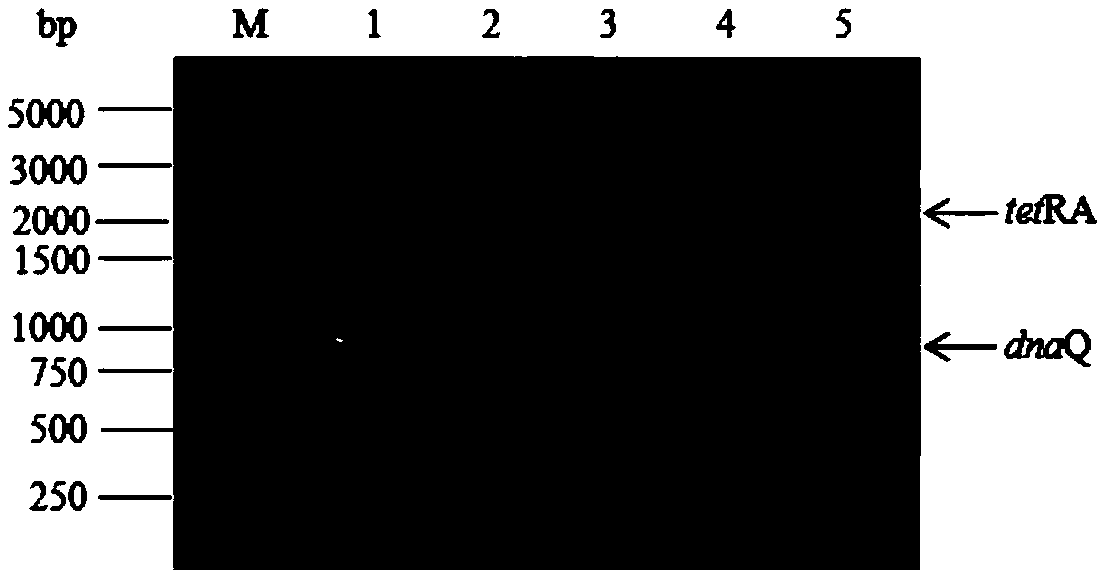
[0035] The features and advantages of the present invention can be further understood through the following detailed description in conjunction with the accompanying drawings. The examples provided are only illustrative of the method of the present invention and do not limit the rest of the present disclosure in any way. [Example 1] The gene dnaQ encoding Escherichia coli DNA polymerase IIIε subunit was knocked out using the λ-Red recombinase method
[0036] (1) Design primers:
[0037] The primers used for homologous recombination are dnaQtetR and dnaQtetA; the design principle is that the 5' end is the homologous region of the dnaQ gene (not underlined), and the 3' end is the homologous sequence of the tetracycline resistance gene tetRA (underlined), recombination The primers used for body detection were EdnaQU and EdnaQD. The sequences of these primers are shown in Table 1 and were synthesized by Beijing Aoke Dingsheng Biotechnology Co., Ltd.
[0038] Table 1 The primers...
Embodiment 2
[0046] [Example 2] Functional analysis of dnaQ mutants
[0047] Pick the wild-type DH5α and [Example 1] the dnaQ mutant identified by PCR with EdnaQU and EdnaQD primers and culture them in 5mL LB medium at 200r / min at 37°C overnight, and then collect the cells by centrifugation, concentrate to 1ml, and gradient Dilute, then take 100 μl and spread on LB solid plate and LB solid plate containing nalidixic acid (the concentration of nalidixic acid is 30ug / ml), after culturing at 37°C for 18 hours, observe the growth of the colonies and record the number of colonies on each plate (CFU). The ratio of the number of colonies on the nalidixic acid plate to the number of colonies on the LB plate was used as the mutation rate of resistance to nalidixic acid, and each sample was repeated three times.
[0048] The wild-type DH5α and the dnaQ mutant of [Example 1] were shaken and cultured in LB medium at 37°C overnight. After the overnight culture solution was serially diluted, the same ...
Embodiment 3
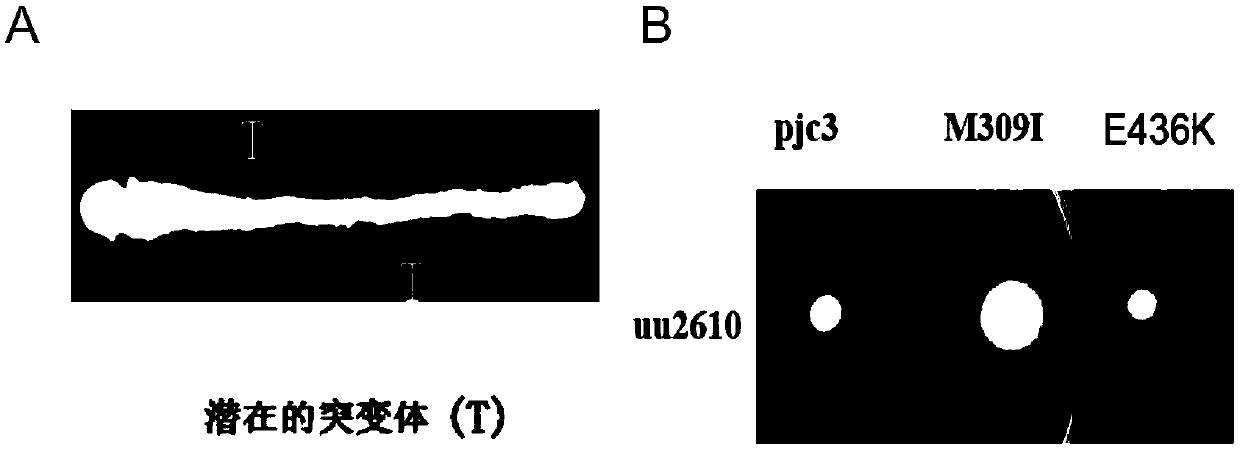
[0054] [Example 3] Mutation test of serine receptor protein gene tsr
[0055] The plasmid pJC3 containing the serine receptor protein gene tsr was transformed into the competent cells of the dnaQ mutant of [Example 1] by heat shock method, plated on an LB plate containing 100ug / ml ampicillin, and overnight at 37°C to cultivate. Pick different colonies and inoculate them into 1ml of LB liquid medium respectively. After overnight shaking culture at 37°C, take 10ul of the culture solution and spread it on a plate containing 30ug / ml nalidixic acid, and culture it overnight at 37°C. Plasmids were extracted from the original colony that produced the largest number of resistant colonies on the nalidixic acid plate, and used as a tsr mutation library.
[0056] The mutant library plasmid was transformed into strain uu2610 (△MCPs△CheRCheB) whose methyl modification system CheR, CheB and all chemotactic receptors were knocked out by heat shock method. Streak the transformed cells on a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com