HRM primers and methods for differentiating Mycoplasma capricosum subsp.
A technology of mycoplasma and goats, applied in the field of rapid diagnosis of veterinary infectious diseases, can solve the problems of HRM primers and methods that have not yet been identified, and achieve rapid cost, strong specificity, and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] 1. Strain
[0022] Mycoplasma capricolum subspecies FJ-GT strain and Mycoplasma capricolum subsp. caprine pneumonia standard strain MccpF38 were all preserved by the Laboratory of Herbivore Diseases, Institute of Animal Husbandry and Veterinary Medicine, Fujian Academy of Agricultural Sciences.
[0023] 2. Primer design and synthesis
[0024] Two pairs of specific primers were designed according to the sequence of Mmc 95010 strain MLC_0560 gene (GenBank accession number FQ377874.1) and the sequence of Mccp F38 strain MCCPF38_00984 gene (GenBank accession number LN515398.1):
[0025] Outer primer SEQ ID No. 5: MFW325: 5'-GAATGTTTAATCGATCAACTGCT-3',
[0026] SEQ ID No. 6: MRW325: 5'-TGCTAATAGTGGTCCTGCTAAGA-3'.
[0027] Inner primer SEQ ID No.7: 107F: 5'-CAATACATCCATTARMACTCTTGA-3',
[0028] SEQ ID No. 8: 107R: 5'-GAATWCAYTCAGGTTGATTATTAGGA-3'.
[0029] 3. The HRM method for identifying Mycoplasma capricolum subsp. capricolum and Mycoplasma capricolum subsp. capricolum...
Embodiment 2
[0038] Example 2 Specificity Verification
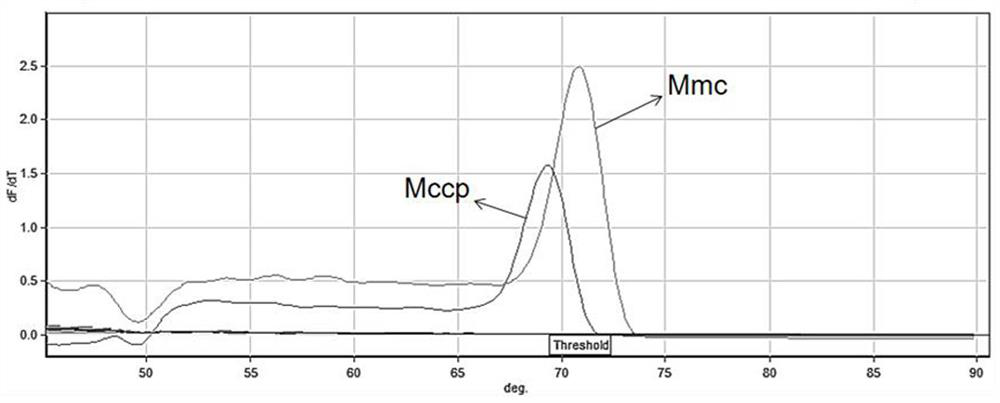
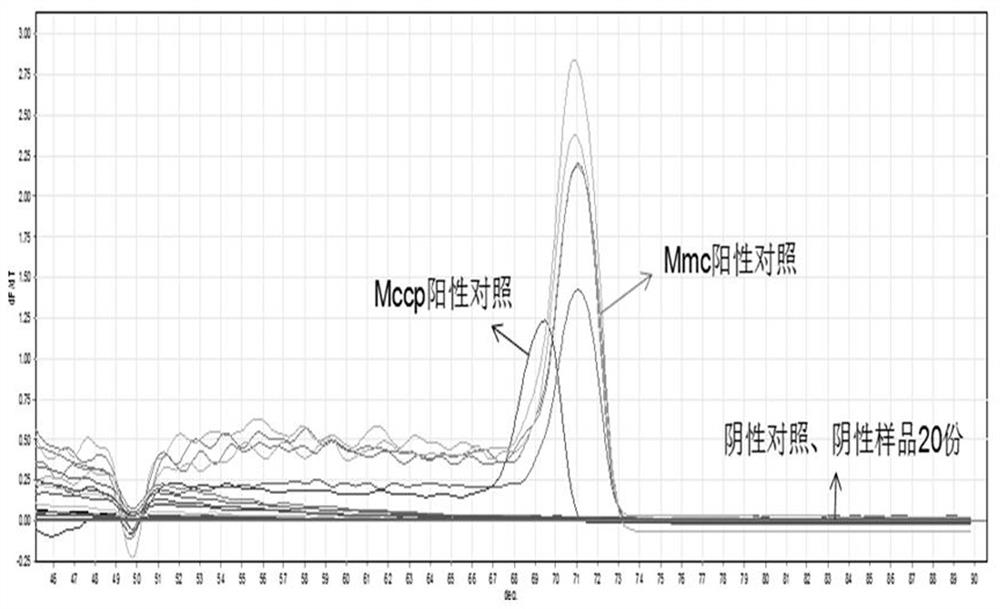
[0039] Use the method of step (1) in Example 1 to extract the genomic DNAs of Mmc, Mccp, Mycoplasma ovine pneumonia (Movi), Orchitis Virus (ORFV), Escherichia coli, Salmonella, Pasteurella, and Mycoplasma arginine, respectively as templates , using HRM method for identification, to verify the specificity of the established HRM method for identification of Mycoplasma capricolum subspecies and Mycoplasma capricolum subspecies caprine pneumonia. see the results figure 1 . figure 1 The results show that the invention can accurately and rapidly identify Mycoplasma capricolum subspecies and Mycoplasma capricolum capitis subspecies, and can be used for clinical diagnosis and epidemiological investigation of Mycoplasma capricolum pneumonia and caprine infectious pleuropneumonia.
Embodiment 3
[0040] Example 3 Repeatability Verification
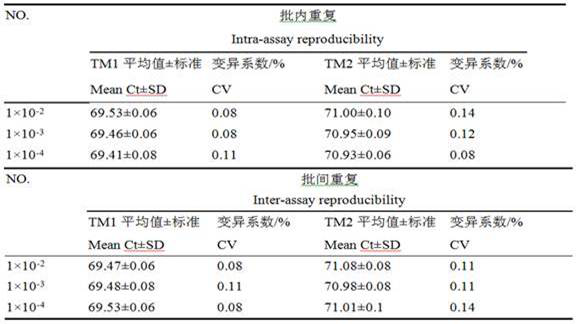
[0041] The Mmc and Mccp genomic DNAs extracted by Ezup column bacterial genomic DNA extraction kit were used as templates, and the extracted templates were amplified by PCR with primers MF-107 and MR-107, and the amplified products were recovered and purified after agarose gel electrophoresis. , connect the purified target gene to PMD-19T cloning vector, transform DH5α competent cells, use TaKaRaMiniBEST Plasmid Purification Kit Ver.4.0 to extract the plasmid for PCR identification and enzyme digestion identification, measure its concentration and calculate the copy number as the HRM detection Standard. The standard was diluted 10-fold as a template for repeatable HRM detection. The intra-batch repeat test is to measure the samples of each concentration 3 times; the inter-batch repeat test is to measure 3 times of the standard substance of 3 dilutions, and the interval of each time is 3d. The HRM reaction system and cycle parameters...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com