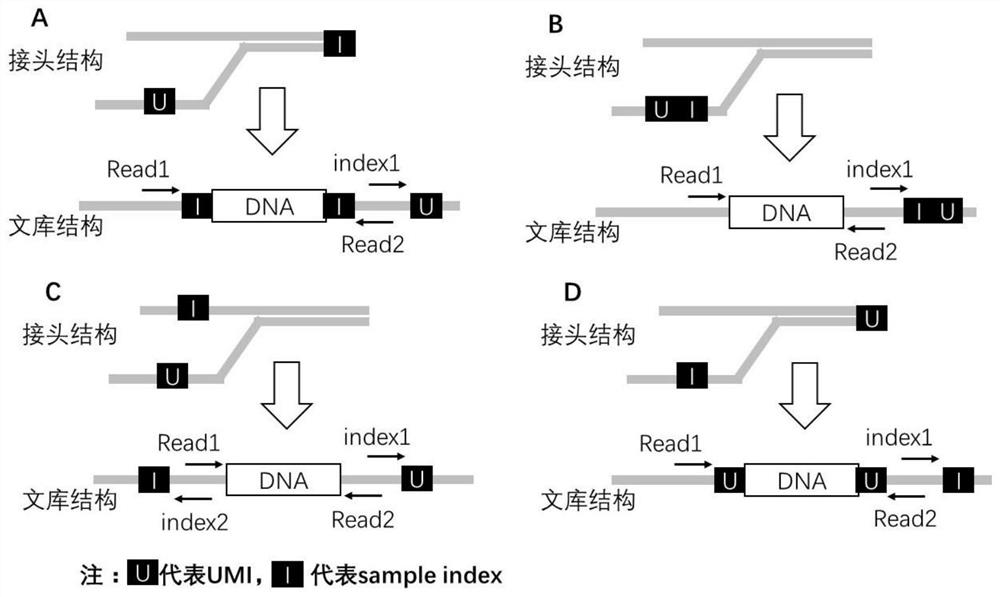
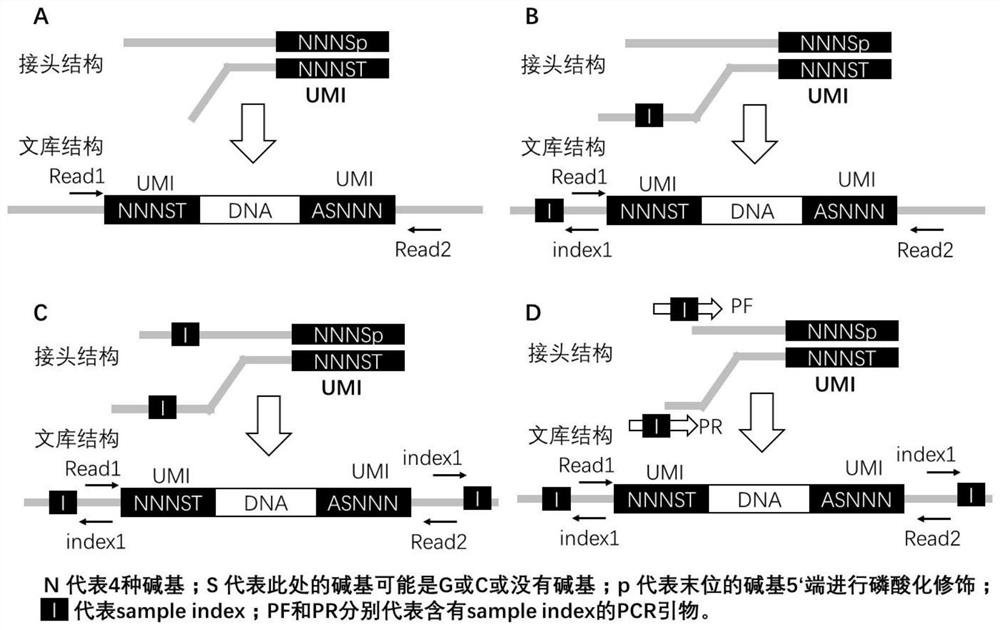
Double-end molecular index adapter, its use and sequencing library with the adapter
A technology of molecular tags and linker sequences, which is applied in the field of sequencing, can solve problems such as interference with high-throughput sequencing technology detection, RNA qualitative errors, cumbersome operations, etc., and achieve the effects of improving sequencing data quality, optimal connection efficiency, and simple preparation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] In this example, the conventional sequencing joints of BGISEQ and MGISEQ series sequencers of BGI were used to design and synthesize double-ended UMI joints, including:
[0066] 64 kinds of short chain linker sequences: 5'-GAACGACATGGCTACGATCCGACTTNNNST-3' (SEQ ID NO: 3), wherein UMI is selected from the above table 1;
[0067] 64 kinds of long-chain linker sequences: 5'-pSNNNAAGTCGGAGGCCAAGCGGTCTTAGGAAGACAA-3' (SEQ ID NO: 4), wherein UMI is selected from Table 1 above.
[0068] In the above sequence, p represents phosphorylation modification, S represents G or C or no base. In this example, the above 128 sequences were synthesized by Beijing Liuhe Huada Gene Technology Co., Ltd., the purification method was PAGE plus, and the order quantity was 5OD.
[0069] The synthetic DNA sequence dry powder was centrifuged at 12000 rpm for 2 min. Dilute the primers to 100 μM with TE buffer, and the TE configuration is shown in Table 2.
[0070] Table 2 TE buffer configuration ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com