Protein synthesis efficiency enhancing RNA element
A protein synthesis and component technology, applied in the biological field, can solve problems such as less translation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0077] In the present invention, the preparation method of the yeast cell extract is not limited, and a preferred preparation method includes the following steps:
[0078] (i) providing yeast cells;
[0079] (ii) washing the yeast cells to obtain washed yeast cells;
[0080] (iii) subjecting the washed yeast cells to destructive treatment to obtain crude yeast extract;
[0081] (iv) performing solid-liquid separation on the crude yeast extract to obtain the liquid part, which is the yeast cell extract.
[0082] In the present invention, the solid-liquid separation method is not particularly limited, and a preferred method is centrifugation.
[0083] In a preferred embodiment, said centrifugation is performed in a liquid state.
[0084] In the present invention, the centrifugation conditions are not particularly limited, and a preferred centrifugation condition is 5000-100000×g, preferably 8000-30000×g.
[0085] In the present invention, the centrifugation time is not parti...
Embodiment 1
[0144] Example 1: Determination of the endogenous IRESs sequence of eukaryotic cells
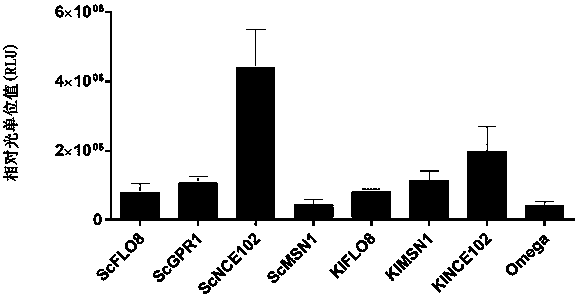
[0145] 1.1 Saccharomyces cerevisiae ( Saccharomyces cerevisiae ) Determination of the sequence of endogenous IRESs: find out the 60 bases upstream of the start codon (ATG) of 9 related genes (Table 1) in the Saccharomyces cerevisiae genome, and name them as the sequences of the corresponding IRESs are ScYMR181c, ScGPR1, ScBOI1, ScFLO8, ScNCE102, ScMSN1, ScGIC1, SceIF4G2 and ScPab1.
[0146] 1.2 Kluyveromyces lactis ( Kluyveromyces lactis ) Determination of the sequence of endogenous IRESs: use Blast to find the homologous genes of 9 genes in 1.1 from the K. lactis genome (Table 1), and then determine the 60 upstream of the start codon (ATG) bases, and as the corresponding IRESs sequences, named KlYMR181c, KlGPR1, KlBOI1, KlFLO8, KlNCE102, KlMSN1, KlGIC1, KleIF4G, and KlPab1, respectively. Since only one homologous gene of SceIF4G2 was found in the K. lactis genome, only 60 bases upstrea...
Embodiment 2
[0149] Embodiment 2: Contain the construction of the in vitro protein synthesis system plasmid of eukaryotic cell endogenous IRESs
[0150] 2.1 Whole-gene synthesis: The sequences of nine IRESs derived from Kluyveromyces lactis were concatenated and synthesized using the method of whole-gene synthesis.
[0151] 2.2 Construction of the plasmid: using a pair of long primers for the 9 IRESs (ScIRESs) derived from Saccharomyces cerevisiae, the sequence of the IRESs was inserted into the existing plasmid Omega-Fluc (SEQ ID NO. 19) by PCR to replace the Omega sequence, respectively The plasmids containing Saccharomyces cerevisiae IRESs were constructed, and their names and sequence numbers are listed in Table 2.
[0152] For the 9 IRESs (KlIRESs) derived from Kluyveromyces lactis, two pairs of primers were used, the primers with the suffixes 1f and 1b were used to amplify the corresponding IRESs fragments from the synthetic DNA fragments respectively, and the primers with the suffix...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com