Malic dehydrogenase PbMDH as well as coded sequence and application thereof
A technology of malate dehydrogenase and nucleotide sequence, which is applied in the field of genetic engineering and enzyme engineering, and can solve the problems of uncommon mining of malate dehydrogenase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Activation and cultivation of embodiment 1 Pseudomonas betel (Pseudomonas beteli)
[0063] Transfer Pseudomonas beteli (Pseudomonas beteli) stored at -80°C into 10 mL of 2216L medium according to 1% inoculum amount, place it in a constant temperature shaker at a temperature of 25°C and a speed of 200 rpm, and activate it for 12 hours . The activated strain was inoculated into 50 mL of 2216L medium at a ratio of 1%, and cultured in a constant temperature shaker at a temperature of 25° C. and a rotation speed of 200 rpm until the OD600 was about 0.8.
Embodiment 2
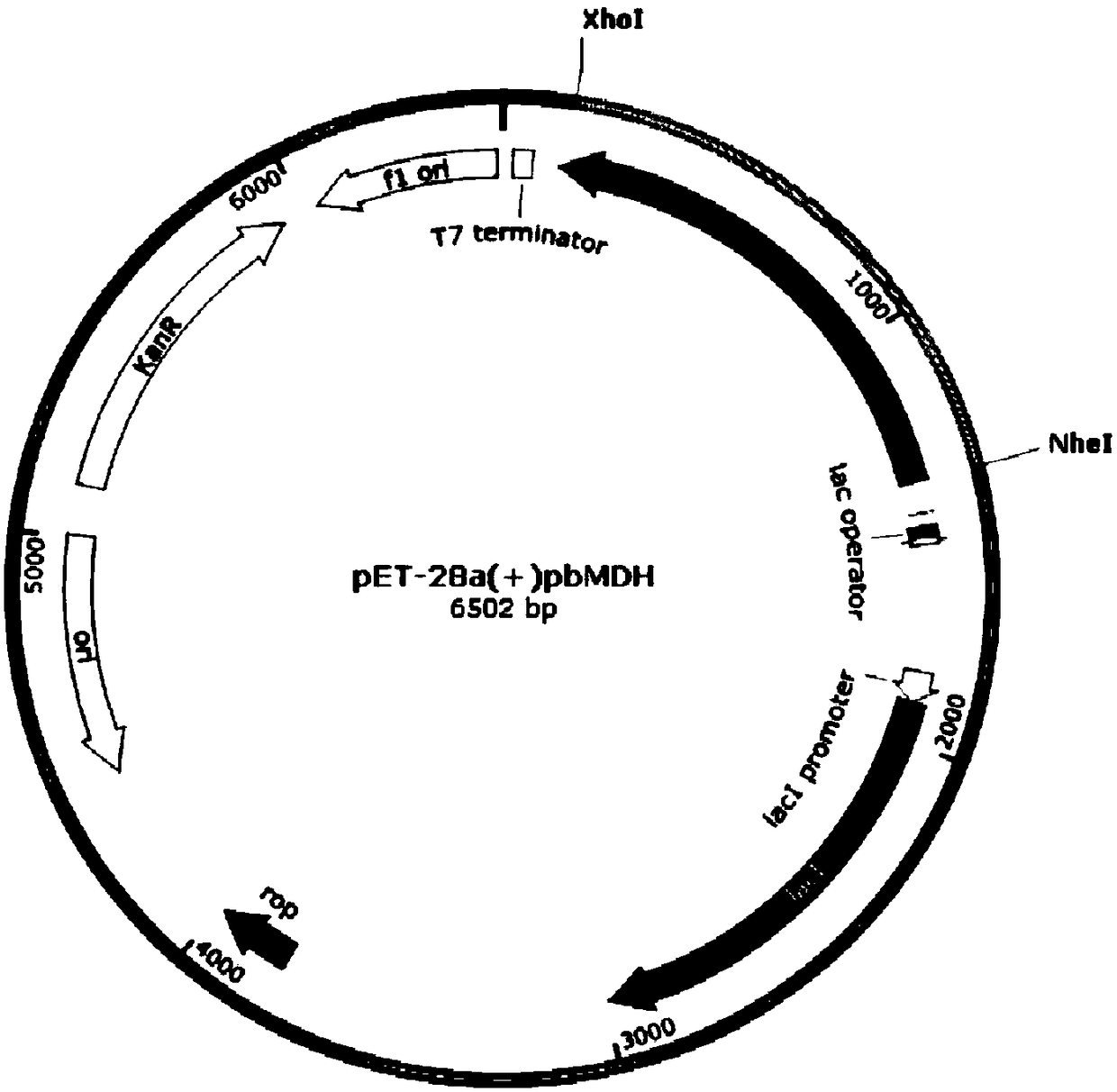
[0064] Example 2 Gene cloning of Pseudomonas beteli malate dehydrogenase PbMDH and construction of recombinant vector
[0065] The total RNA of Pseudomonas beteli was extracted using the genome extraction kit produced by Takara Company, and then cDNA was synthesized by reverse transcription, and 1 μL of cDNA was used as a template for polymerase chain reaction.
[0066] According to the nucleotide sequence of SEQ ID NO.2, a pair of primers are designed for PCR amplification, and the primers are designed as follows:
[0067] Upstream primer PbMDHF1 (SEQ ID No.9): 5'-GGAATTC CATATG TCHGATYTYAAAACYGCYG-3';
[0068] Downstream primer PbMDHR1 (SEQ ID No.10): 5'-CCG CTCGAG TYARCCSTTGAASACRTCRTC-3';
[0069] Wherein, YRS represents a degenerate base, specifically Y=C / T, R=A / G, S=G / C, and the underlined sequence is the restriction endonuclease site NdeI and XhoI;
[0070] Using the extracted Pseudomonas beteli genome cDNA as a template, the PCR amplification system is as follows...
Embodiment 3
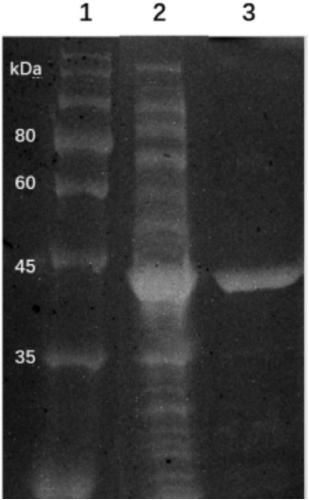
[0081] The construction of embodiment 3 recombinant escherichia coli
[0082] The specific steps of transferring the successfully connected recombinant plasmid into Escherichia coli are as follows:
[0083] (1) Take 100 μL of competent cells and place them in an ice bath to melt for 10 minutes;
[0084] (2) Add 0.1ng-10ng (3μL-10μL) of the transformed recombinant plasmid to the competent cells, mix gently and place on ice for 30min;
[0085] (3) Place the above bacterial solution at 45°C for heat shock for 45 seconds, then quickly put it in an ice bath for 1-2 minutes;
[0086] (4) Add 890 μL fresh LB medium preheated at 37°C, and incubate at 37°C, 200 rpm for 1 hour;
[0087] (5) Spread 100-200 μL of the bacterial solution evenly on an LB plate containing kanamycin (100 μg / mL) with a coating stick, and culture overnight at 37°C;
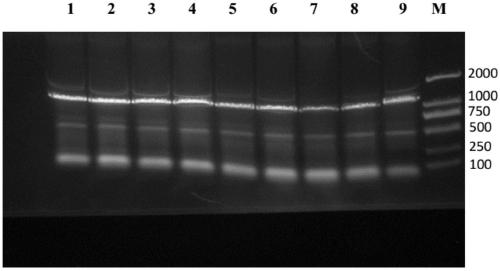
[0088] Pick the transformants on the plate for colony PCR, screen positive clones, and the PCR results are as follows: figure 1 As shown, the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com