Detection method of invasive fungus infection, detection kit and application
A technology of invasive fungi and a detection method, applied in the field of detection kits, to achieve the effects of avoiding the possibility of contamination, saving manpower, and eliminating misdiagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
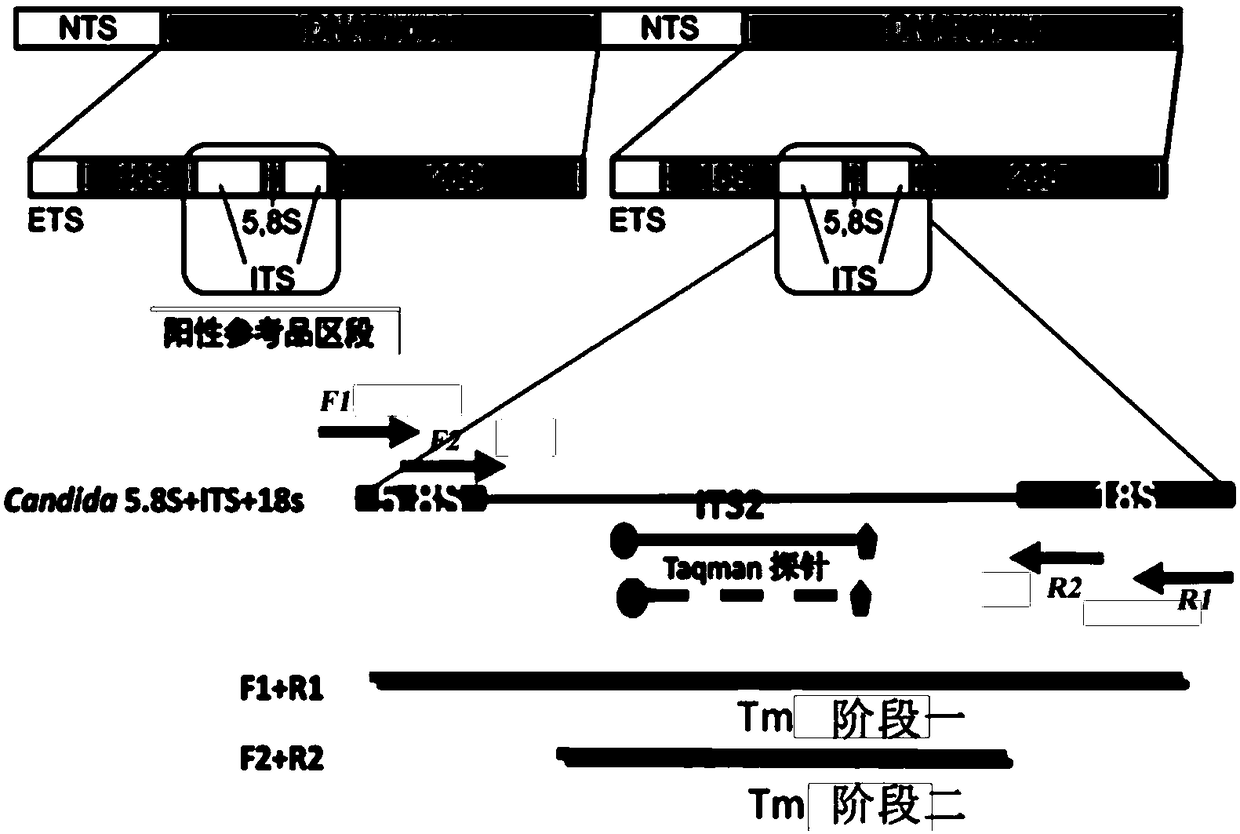
[0107] Verify the method of the present invention with positive plasmid sample
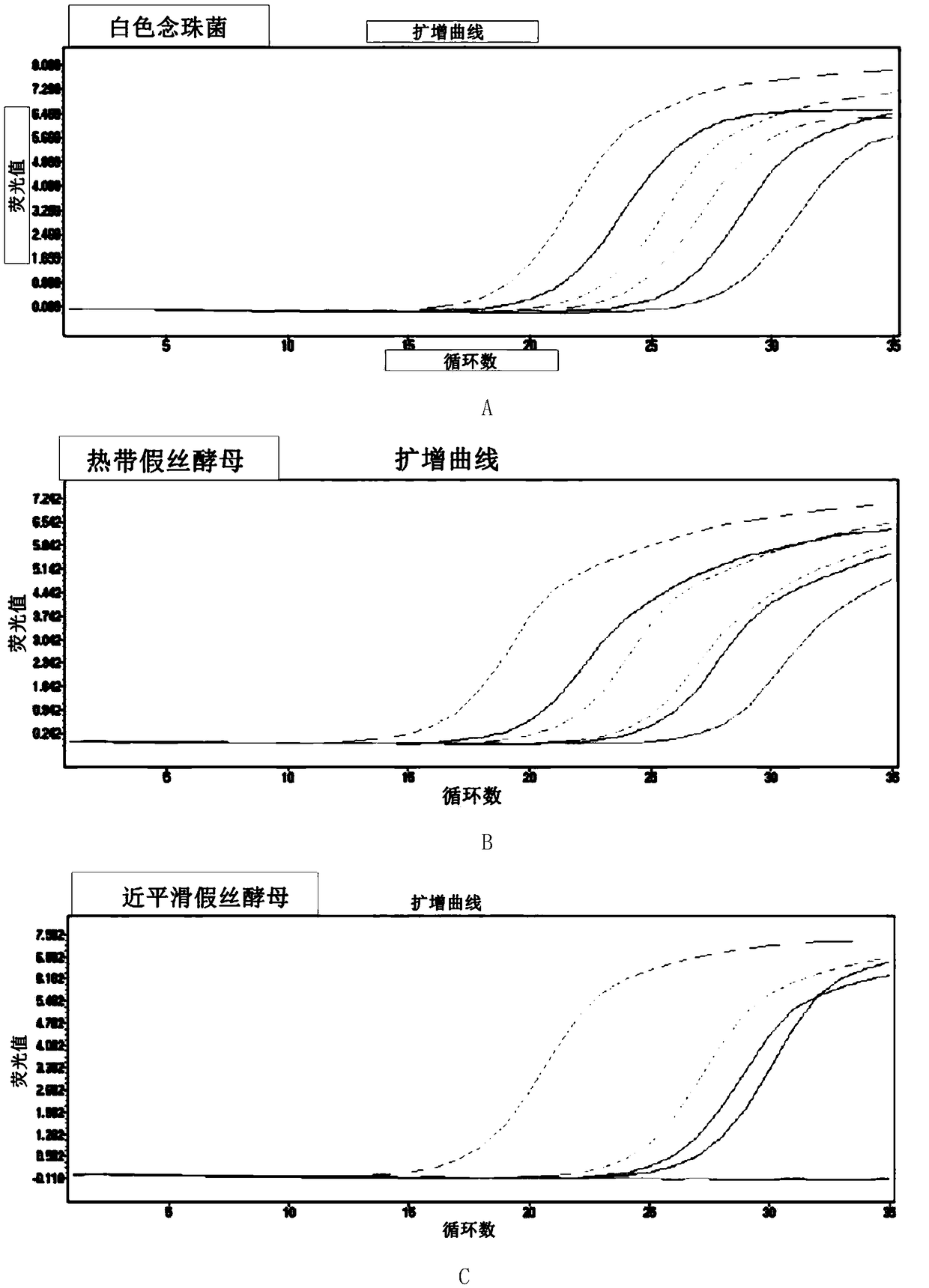
[0108]In this example, the method of the present invention is used to carry out PCR detection on the plasmids of each positive bacterial species, and it is verified that the reaction system of the method of the present invention can be used to specifically identify Candida albicans, Candida tropicalis, and Candida smoothie at one time through multiple reactions in a single tube. Yeast or Candida krusei, Candida glabrata, Aspergillus fumigatus without cross-reaction. The positive plasmids of the above-mentioned bacterial species were included in each reaction, and the positive plasmids of each bacterial species were prepared individually or in combination into samples to be tested (plasmid concentration was 10 2 copies / μL).
Embodiment approach
[0110] 1. Prepare positive plasmids for strains
[0111] 1.1. Primer design: refer to the reported rDNA sequences of each strain (including 5.8s, 18s, 28s and ITS regions) to design and construct the primer sequences of the positive plasmids of each strain:
[0112] Candida albicans:
[0113] The 5'-3' sequence of the upstream primer: GGTGTTGAGCAATACGACTTGG (SEQ ID NO.15)
[0114] Downstream primer 5'-3' sequence: AGACCTAAGCCATTGTCAAAGC (SEQ ID NO.16)
[0115] Candida tropicalis:
[0116] Upstream primer 5'-3' sequence: TGGTATTCCAAAGGGCATGC (SEQ ID NO.17)
[0117] Downstream primer 5'-3' sequence: CCACGTTAAATTCTTTCAAAACAAA (SEQ ID NO.18)
[0118] Candida smoothie:
[0119] Upstream primer 5'-3' sequence: ATTGCGCCCTTAGGGCATG (SEQ ID NO.19)
[0120] Downstream primer 5'-3' sequence: TCCATTAGTTTATACTCCGCCTT (SEQ ID NO.20)
[0121] Candida krusei:
[0122] Upstream primer 5'-3' sequence: CTGTTTGAGCGTCGTTTCCA (SEQ ID NO.21)
[0123] Downstream primer 5'-3' sequence: TCCTAC...
Embodiment 2
[0149] Species identification testing using in vitro culture models infected with known species
[0150] In this example, the culture supernatant sample of in vitro simulation (PBMC) of certain fungal strains was used to test whether the PCR system of the present invention can use free fungal DNA to distinguish strains without being affected by non-detection target strains. Through this example, it is further confirmed that the kit of the present invention is suitable for distinguishing and identifying fungal species based on fungal free DNA fragments, and shows good system specificity and repeatability in the detection of in vitro simulated infection samples, without affected by other interfering factors.
[0151] 1. Implementation method:
[0152] 1. For the culture medium of known strains, take the supernatant after centrifugation, and extract DNA with Qiagen or Desay Diagnostic Systems (Shanghai) Co., Ltd. Blood DNA Extraction Kit.
[0153] 2. The real-time PCR reaction ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com