Lactic acid-utilizing bacteria genetically modified to secrete polysaccharide-degrading enzymes
A polysaccharide degrading enzyme and genetic modification technology, applied in the field of lactic acid-utilizing bacteria that are genetically modified to secrete polysaccharide degrading enzymes, can solve the problems of high cost, limited carbohydrate source, low carbon-lactic acid conversion rate, etc., and achieve cost efficiency High, effective mixing of waste, pollution risk reduction effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0165] Preparation of separated lactic acid enantiomers
[0166] In some embodiments, the reducing sugars of the organic waste (those reducing sugars originally found in the waste and those released by the action of polysaccharide degrading enzymes) can be fermented into lactic acid by microorganisms producing LA. In order to produce only one isolated lactic acid enantiomer, the LA-producing microorganism used produces only one of D-lactic acid and L-lactic acid enantiomer. The LA-producing microorganism can naturally produce only one enantiomer, or can be genetically modified to produce only one enantiomer, for example, by knocking out one or more enzymes involved in the synthesis of unwanted enantiomers.
[0167] In some embodiments, prior to lactic acid fermentation, the LA-utilizing bacteria are inactivated to avoid consumption of lactic acid produced during fermentation. For example, inactivation can be performed by increasing the temperature or changing the pH to a temperat...
Embodiment 1
[0211] Example 1-Construction of suitable vectors for genetically modified bacteria
[0212] Construction of Escherichia coli-Propionibacterium freudenreichii shuttle vector:
[0213] The constructed vector can be used in both Escherichia coli and Propionibacterium freudenreichii.
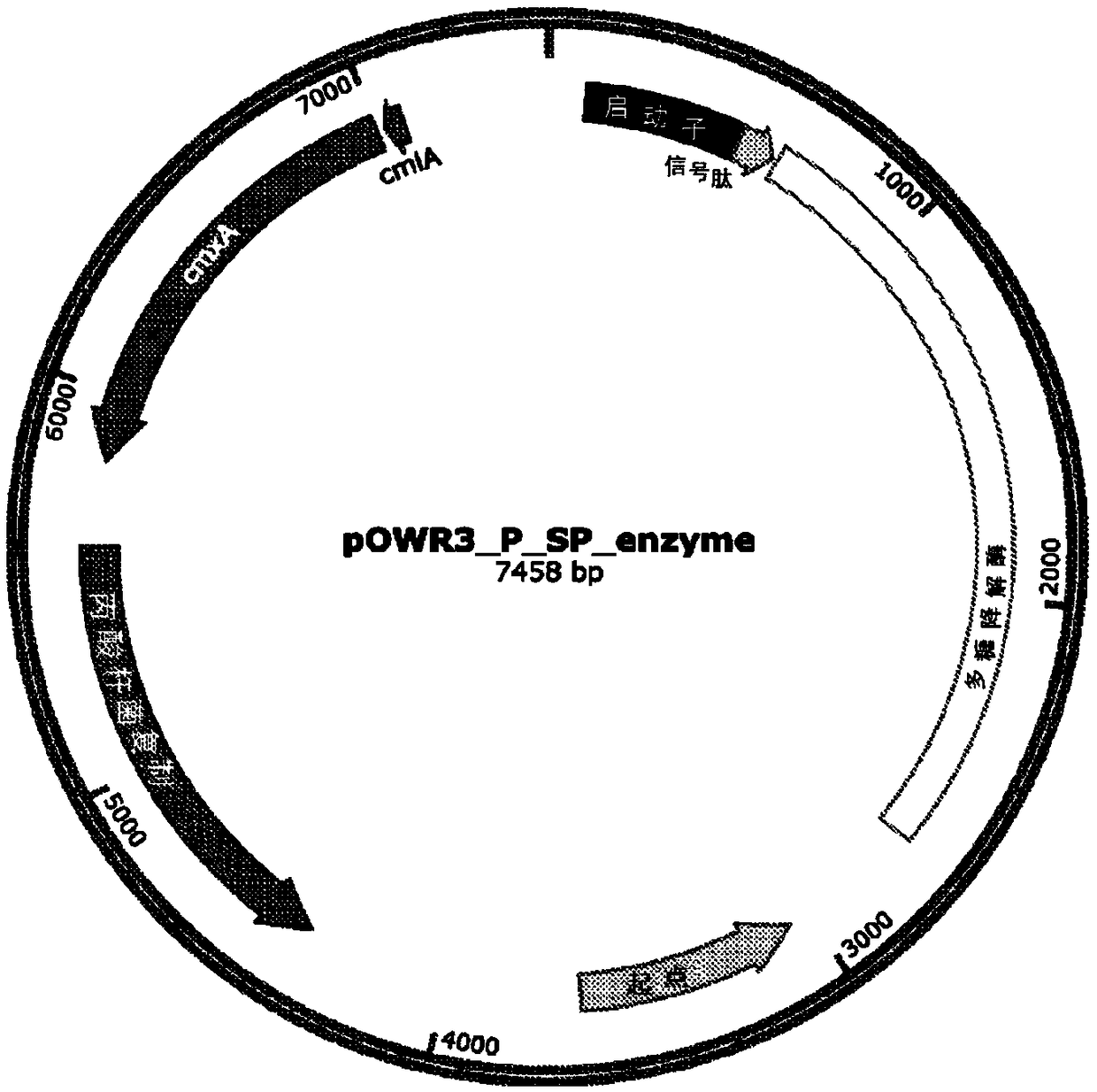
[0214] Prepare the pOWR3 vector as follows (the schematic diagram is shown in Figure 2A Middle): Propionibacterium replication protein sequence (SEQ ID NO.84) and chloramphenicol resistance gene (cmx (A) (SEQ ID NO: 85) and cml (from Corynebacterium striatum pT10 plasmid) A) (SEQ ID NO: 86)) was synthesized in tandem into a vector containing the E. coli origin of replication (ori) and the ampicillin resistance cassette (AmpR). This results in a ~5.6 kbp plasmid that can replicate in both E. coli and P. freudenreichii. The vector also includes the PB origin of replication sequence. The resulting pOWR3 vector is smaller than other known P. freudenreichii expression vectors (5.6 kbp vs. 6.2-8 kbp), whi...
Embodiment 2
[0219] Example 2—Construction of suitable expression cassettes and expression vectors for genetic modification of bacteria to express and secrete polysaccharides Degrading enzyme
[0220] Listed below are exemplary polysaccharide degrading enzymes (glucoamylase, cellulase, and hemicellulase (xylanase), signal peptides, and promoters used to generate expression cassettes used in expression vectors.
[0221] The following exemplary enzymes listed in Table 1 below are used for expression cassettes and expression vectors for genetically modified bacteria. Enzyme sequences were obtained from different sources (biological). The original signal peptide of each listed enzyme was removed and replaced with an exogenous (different) signal peptide.
[0222] Table 1-Exemplary polysaccharide degrading enzymes
[0223]
[0224]
[0225] The following promoter regions are used to construct various expression cassettes and expression vectors:
[0226] -PFREUD_04850 promoter from Propionibacterium fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com