Preparation method for dual-loaded targeted nano platform of dendrimer based on fluorescent carbon dot modification
A technology of dendritic macromolecules and fluorescent carbon dots, which is applied in the direction of pharmaceutical formulations, preparations for in vivo tests, and medical preparations of non-active ingredients. It can solve problems such as simultaneous connection, achieve reduced inhibitory effects, and good water solubility. and biocompatibility, low-cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] (1) Add RGD polypeptide solution (6.85 mg, 5 mL DMSO) dropwise to MAL-PEG-COOH (19.83 mg, 5 mL DMSO), stir for 3 days, and then use a dialysis bag with a molecular weight cut-off of 5000 to dialyze the aqueous solution ( 2L / time, 3 times / day), freeze-dry to get RGD-PEG-COOH.
[0051] (2) Add EDC solution (18.21mg, 5mLDMSO) to RGD-PEG-COOH solution (18.33mg, 5mL DMSO), stir at room temperature for 30 minutes, then add NHS solution (9.43mg, 3mL DMSO) and continue stirring for 3 hours to obtain Mixed solution; add the mixed solution dropwise to the fifth generation polyamidoamine dendrimer G5-NH 2 solution (38.34mg, 10mL DMSO), the stirring reaction time was 3 days, and then the aqueous solution was dialyzed (2L / time, 3 times / day) using a dialysis bag with a molecular weight cut-off MWCO of 5000, and freeze-dried to obtain G5-PEG- RGD.
[0052] (3) Add CDI solution (25.23mg, 10mL DMSO) to TPGS solution (10.10mg, 4mL DMSO), stir at room temperature for 6 hours, then add G...
Embodiment 2
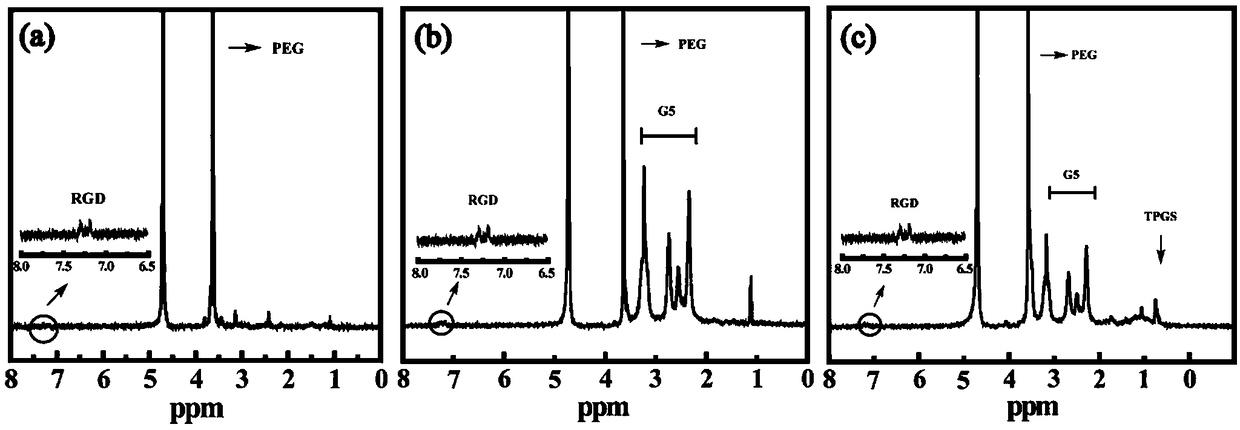
[0057] Take the RGD-PEG-COOH prepared in Example 1, G5-PEG-RGD and G5-RGD-TPGS were dissolved in D 2 In O, use Bruker400MHz nuclear magnetic resonance instrument to carry out hydrogen spectrum test. see figure 2 a, In the H NMR spectrum of RGD-PEG-COOH, 3.6ppm is the characteristic proton peak of MAL-PEG-COOH, and 7.15-7.25 is the characteristic proton peak of the benzene ring on RGD. According to the ratio of their integral areas, calculate It is found that 0.89 RGD molecules are linked to each PEG (the molar ratio of the feed is MAL-PEG-COOH:RGD=1:1). see figure 2 b, 2.0-3.6ppm is dendrimer G5.NH 2 The methylene proton peaks, according to the ratio of their integral areas, calculate each G5.NH 2 3.86 PEG-RGD molecules are linked on the above (the molar ratio of the feed is G5.NH2:PEG-RGD=1:5). see figure 2 c, 0.7ppm is the methyl proton peak of TPGS, according to the ratio of their integrated areas, calculates that 9.7 TPGS molecules are linked on each G5-RGD molecu...
Embodiment 3
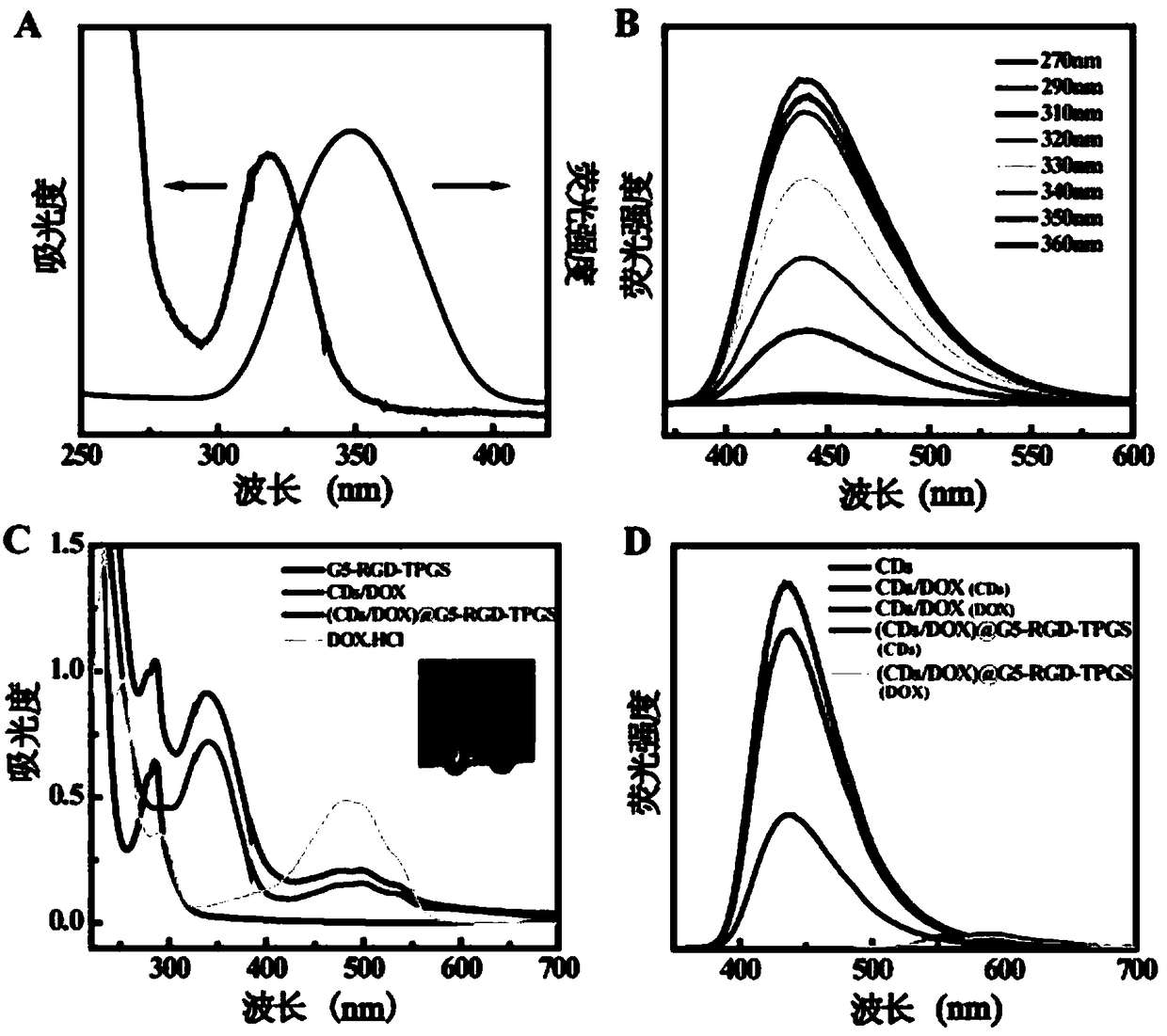
[0059] Dissolve the (CDs-DOX)@G5-RGD-TPGS prepared in Example 1 with pH=7.4 and pH=5.5 buffers respectively to a solution with a concentration of 1mg / mL, take 1mL into a dialysis bag for fixation, and place Place in a container containing 9mL of buffer solution of different pH and shake in a shaker at 37°C. Samples were taken at different time points. Take 1mL of the liquid outside the dialysis bag each time, measure its absorbance at 480nm, and then add 1mL of the corresponding buffer solution to the outside of the dialysis bag. This method was used to obtain the release curves of DOX released from (CDs-DOX)@G5-RGD-TPGS under different pH conditions in vitro. see Figure 4 B, In 70 hours, the release amount of DOX was 48% in a weakly acidic environment (pH=5.5), which was greater than 29.5% in a physiological environment (pH=7.4). It shows that this drug-loading system has a certain pH responsiveness, and the release rate in the weakly acidic environment similar to tumor t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com