Method for visually detecting genetically modified foods
A transgenic and food technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of high detection limit, small detection range, insufficient accuracy, etc., the requirements for meeting the reaction conditions are not high, and the reaction process simple effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
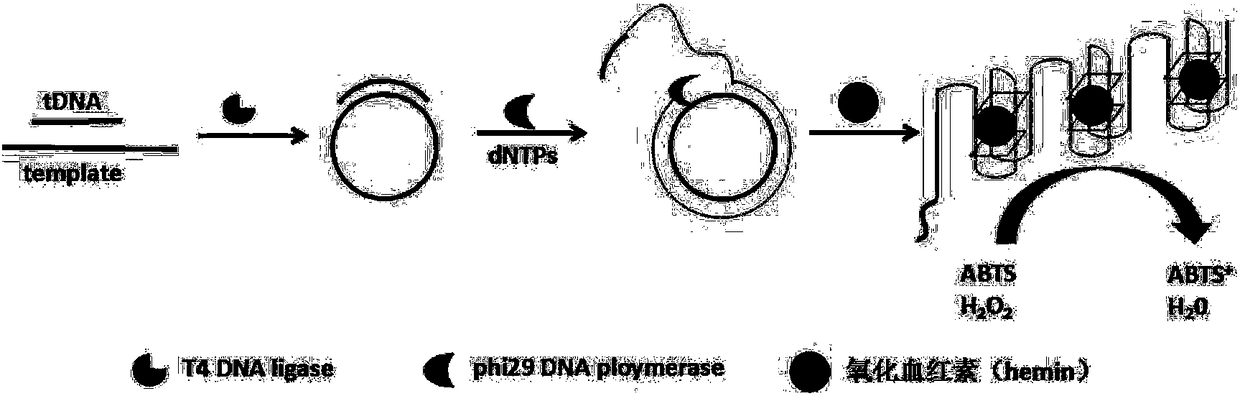
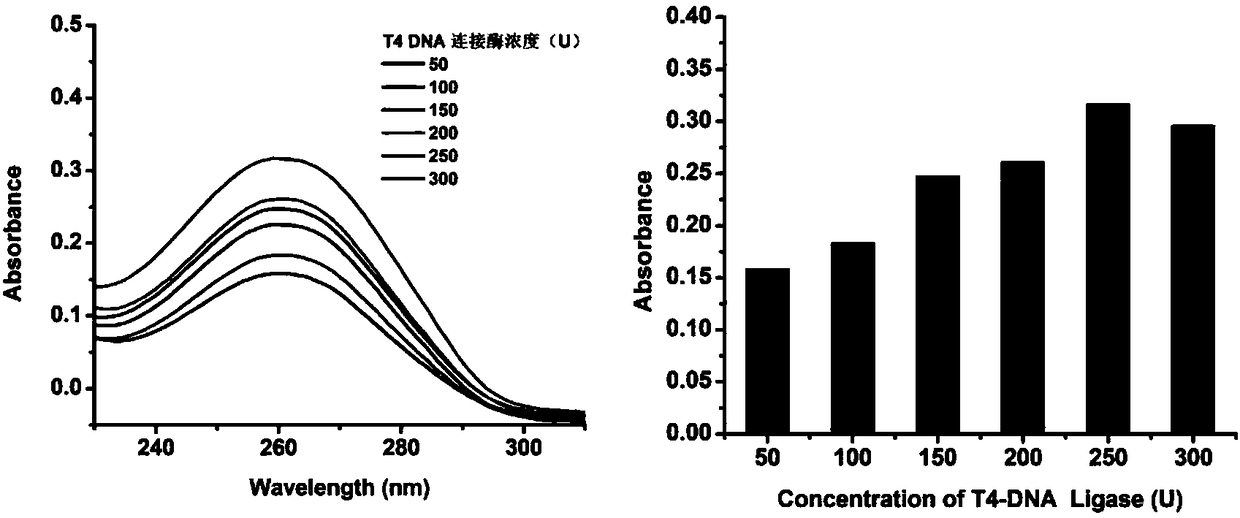
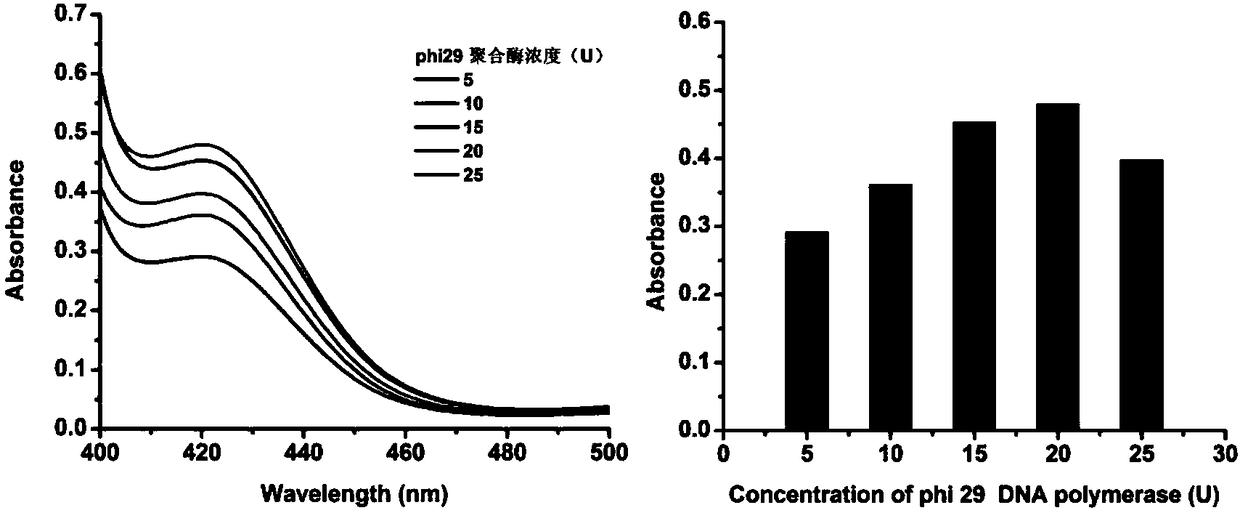
[0050] The principle of visual detection of specific sequence of transgenic soybean by using the RCA synthetic DNase triggered by the target object is as follows: figure 1 shown. First, genomic DNA in transgenic soybeans was extracted using SDS. Design a linear DNA template composed of 42 bases and phosphorylated at the 5' end. The base sequences at both ends are completely complementary to the target gene, and the middle segment contains a base sequence complementary to the G-quadruplex nucleotide sequence . Only in the presence of the CaMV 35S promoter nucleic acid probe, the linear template can be catalyzed by T4-DNA ligase to connect the two ends to form a circular template, and then the RCA reaction is performed under the catalysis of phi29 polymerase to generate a series of circular templates. Complementary long single-stranded amplification products. The amplified product combines metal ions and oxyheme to form DNase, which catalyzes the colorless H 2 o 2 -ABTS rea...
Embodiment 2
[0062] Example 2: Application of the visual detection method of CaMV 35S in the detection of transgenic soybean content
[0063] DNA extraction
[0064] Take 0.1 g of the actual sample in a 5.0 mL centrifuge tube, add 1.0 mL of extraction / lysis buffer, mix well, centrifuge at 12000 rpm for 15 min, and pipette the supernatant into a new centrifuge tube. Add 1 times the volume of Tris-saturated balanced phenol, shake gently and mix well, then centrifuge at 12000rpm for 10min, and take the upper aqueous phase to a new centrifuge tube. Add 1 times the amount of equilibrated phenol-chloroform-isoamyl alcohol (25:24:1) to it, shake gently to mix, centrifuge at 12000 rpm for 10 min, and suck the upper aqueous phase into a new centrifuge tube. Repeat this step until the interphase interface is clean. Then add 1 volume of chloroform-isoamyl alcohol solution, shake gently to mix, centrifuge at 12000rpm for 10min, and draw the upper aqueous phase to a new centrifuge tube again. Repeat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com