Identification method and application of coding protein Pm_MF
An identification method, a protein-encoding technology, which is applied in applications, biochemical equipment and methods, and microbial determination/inspection, etc., can solve problems such as difficulty in functional identification, lack of theoretical support and technical support, and no clear and suitable selection of genes. To achieve the effect of enriching microbial salt-alkali tolerance gene resources and improving salt-alkali tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
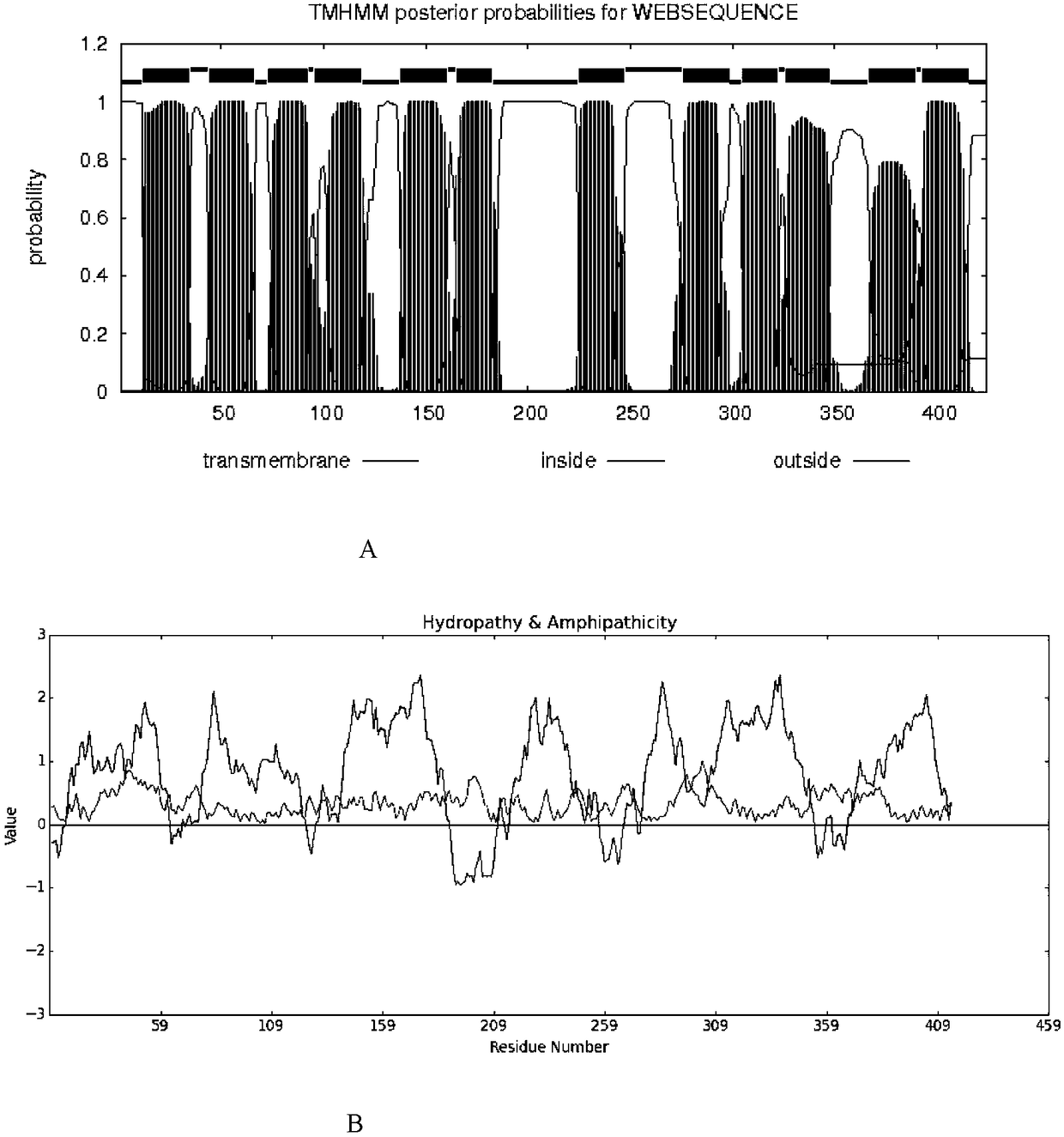
[0127] Bioinformatics analysis of Pm_mf gene and protein Pm_MF, such as figure 1 shown.
[0128] Through the screening technology combining gene library and gene function complementation, a gene fragment related to salt-alkali tolerance was obtained from Zoonococcus littoralis, and the gene fragment related to salt-alkali tolerance contained a complete open reading frame ORF with a length of 1272bp , the nucleotide sequence of ORF is shown in SEQ ID NO.1; sequence analysis shows that it shows the highest homology with a putative MF (majorfacilitator) family transporter, which is 98%, so it is speculated that this gene also encodes A protein of the MF (major facilitator) family, so the gene was named Pm_mf.
[0129] The present invention uses online prediction websites http: / / www.cbs.dtu.dk / services / TMHMM-2.0 / and http: / / web.expasy.org / protscale / to carry out bioinformatics analysis on the protein sequence encoded by Pm_mf gene, It was found that the protein Pm_MF encoded by...
Embodiment 2
[0132] The phylogenetic tree of Pm_MF and the identified Na+ / H+ antiporters and the homology comparison with homologues,
[0133] According to the predicted MF (major facilitator) family location, the present invention establishes a phylogenetic evolutionary tree of family homologues and identified Na+ / H+ antiporters, and the phylogenetic tree shows that the MF (majorfacilitator) family proteins form an independent branch, with a threshold of 99 %, but far from the identified Na+ / H+ antiporter or other protein phylogenetic relationship with Na+ / H+ antiporter activity, so it is considered that Pm_mf encodes a new Na+ / H+ antiporter. In the present invention, the homology comparison of homologs is carried out at the same time, and the results show a high degree of homology, and highly conserved amino acid residues are obtained, wherein the negatively charged amino acid residues include D67, D127, E187, D223, E341; Positive amino acid residues include R71; polar amino acid residue...
Embodiment 3
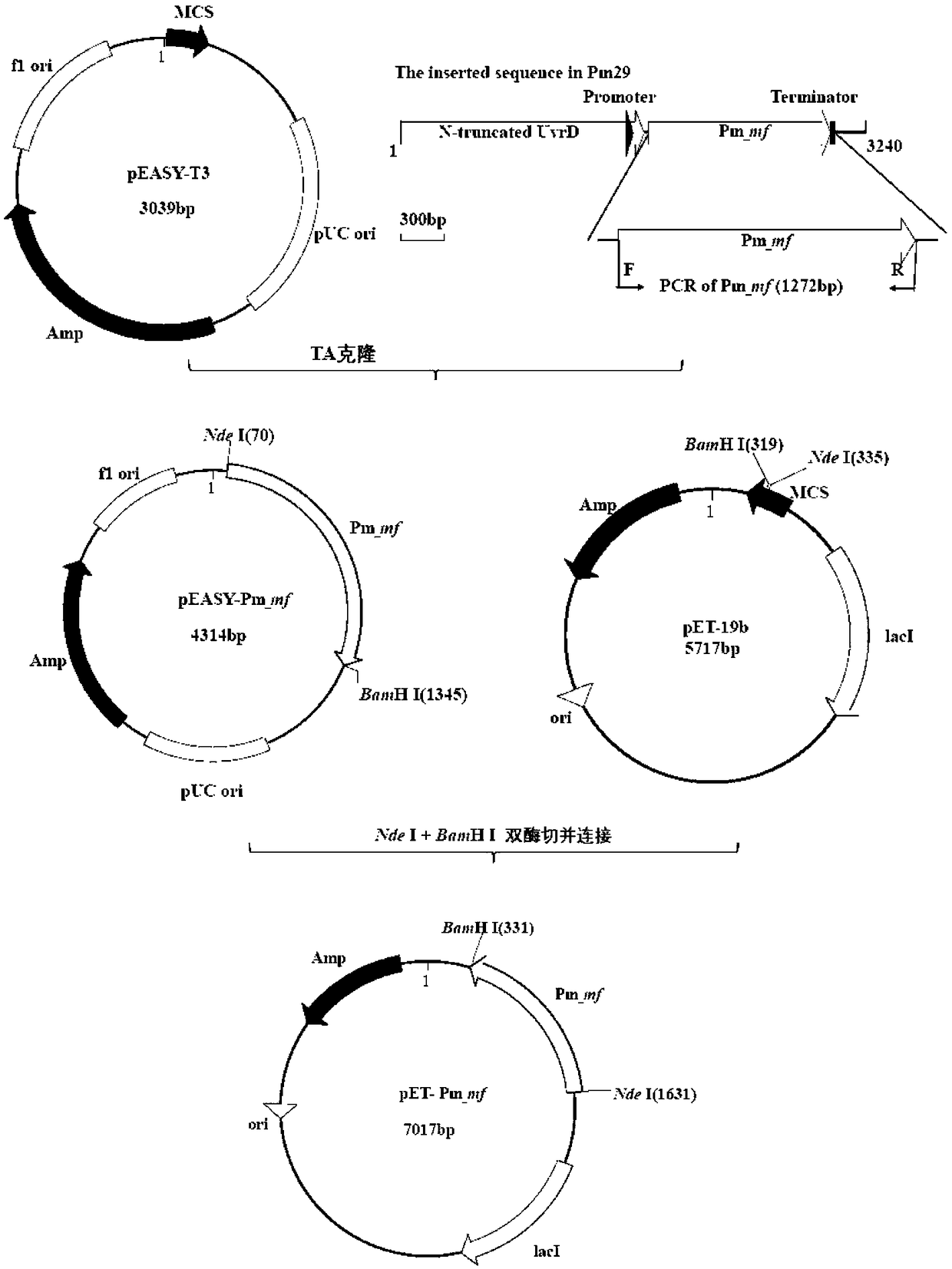
[0135] The construction process of pET-Pm_mf prokaryotic expression vector; as figure 2 shown;
[0136] According to the Pm_mf gene sequence, first use Primer5 software to design SEQID NO.3: Pm_mf-F; SEQID NO.4: Pm_mf-R specific primers (as shown in Table 1), and introduce Nde I at the 5' and 3' ends of the gene respectively , BamH I two enzyme cutting sites, then carry out the amplification of Pm_mf (as shown in table 2), expect to amplify the intermediate sequence fragment length about 1272bp (as shown in Table 2) figure 2 -A shown).
[0137] Table 1 Pm_mf gene expression vector construction primer sequence
[0138]
[0139] Table 2 PCR amplification reaction
[0140]
[0141] Add "A" to the end of the above PCR product and connect it into the pEASY-T3 vector, then transform it into Trans1-T1 competent cells, and perform blue-white screening to select subclones. Recombinant plasmid pEASY-Pm_mf was treated with Nde I and BamH I two kinds of endonucleases, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com