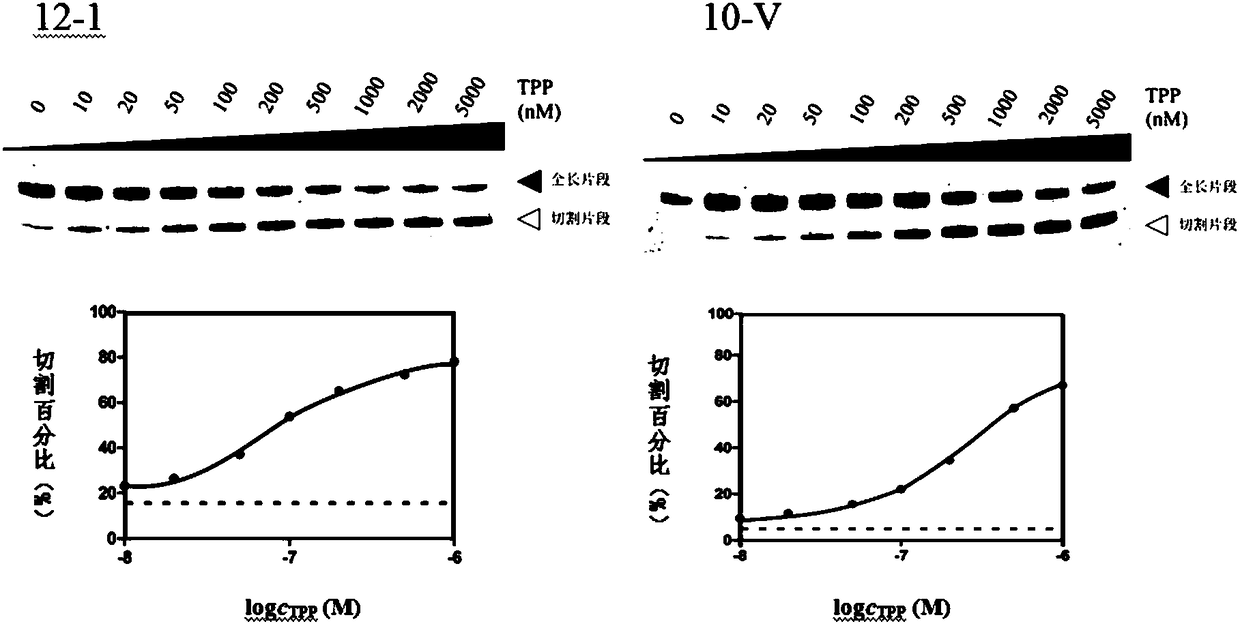
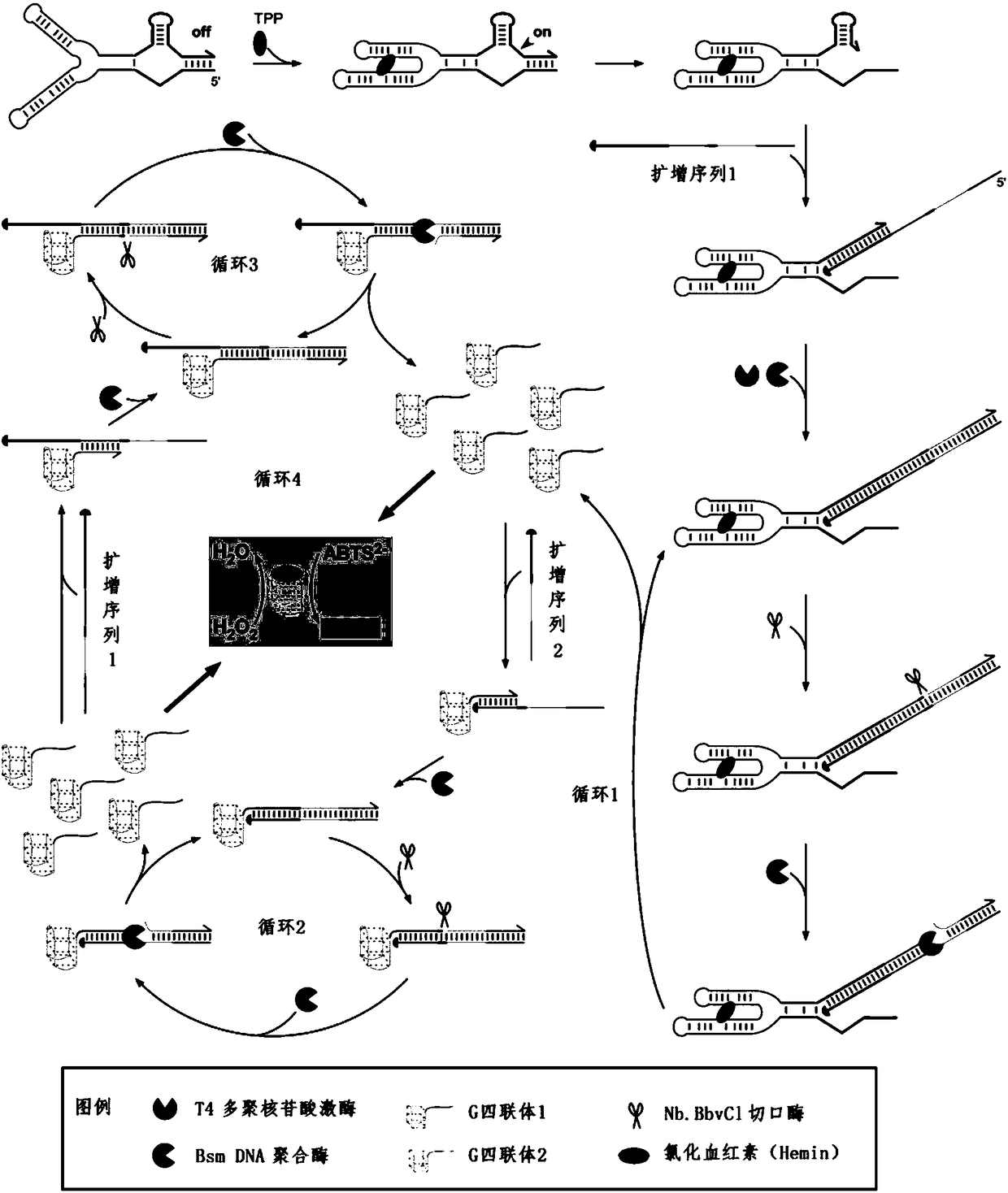
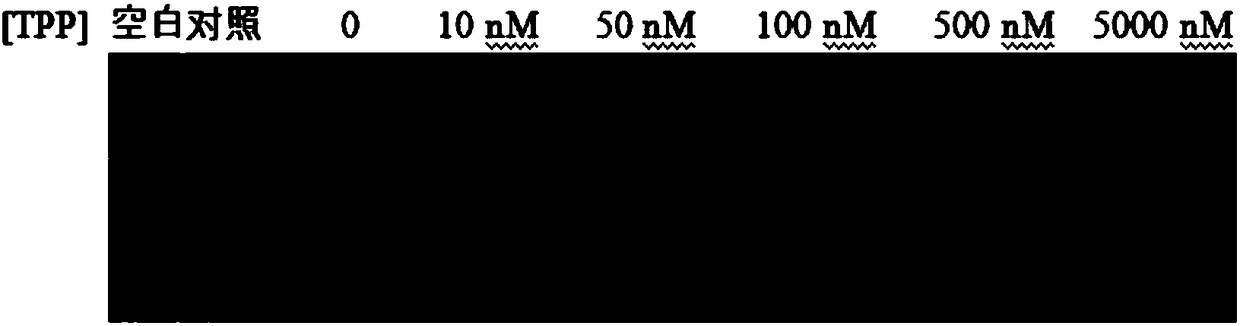
RNA sensor for detecting thiamine pyrophosphate and application thereof
A technology of pyrophosphate and thiamine, applied in the field of RNA sensors, can solve the problems of cumbersome, high operation requirements, and false positive signals, and achieve the effects of short detection cycle, improved detection limit, and convenient sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] The construction of embodiment 1 DNA library
[0055] DNA library construction, such as figure 2 As shown, the DNA sequence was synthesized, purified by 8% denaturing polyacrylamide gel electrophoresis (Polyacrylamide Gel Electrophoresis, PAGE), and the target sequence was recovered with a fluorescence imager and a rubber tapping instrument. Due to technical limitations, the yield of in vitro synthesis of DNA double strands exceeding 100bp is not high, so the present invention uses two segments of single-stranded DNA to extend double-stranded DNA templates in vitro through Polymerase Chain Reaction (PCR), namely Primer 1 (SEQ ID NO: 9) and Primer 2 (SEQ ID NO: 10) are paired with complementary sequences at the ends, and are mutually extended as templates to establish a double-stranded DNA library (random bases are represented by N).
[0056] PCR reaction system:
[0057]
[0058] PCR reaction conditions:
[0059] Pre-denaturation at 95°C for 30s, followed by each...
Embodiment 2
[0064] The construction of embodiment 2 RNA library
[0065] RNA library construction, such as figure 1 As indicated, the above DNA library was transcribed in vitro and incubated at 37°C for 2-3 hours. The transcribed RNA was separated by 8% denaturing PAGE to obtain a sequence that has no cleavage activity in the absence of thiamine pyrophosphate, and the background signal was removed. That is, the RNA library required for screening is constructed, containing 10 random bases.
[0066] Transcription system:
[0067]
Embodiment 3
[0068] Example 3 In vitro screening
[0069] In vitro screening, such as figure 2 shown. The invention adopts the ligand system evolution technology of exponential enrichment, and is divided into steps of pre-screening, screening, reverse transcription, PCR amplification, transcription and the like in vitro. Clones were sequenced after 12 rounds of selection. Specific screening steps include:
[0070] 1. Pre-screening
[0071] Recover the target RNA separated by electrophoresis after transcription, add 50 μl of buffer 1, heat to 70°C for 3-5 minutes, then return to room temperature, add an equal volume of buffer 2 to provide Mg necessary for the enzyme digestion reaction 2+ , allowing the cleavage of sequences that can self-cleavage in the absence of TPP. Perform 8% denaturing PAGE, recover the RNA that has no self-cleavage activity in the absence of TPP, that is, the complete sequence fragment, and elute overnight.
[0072]
[0073] 2. Screening
[0074] After the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com