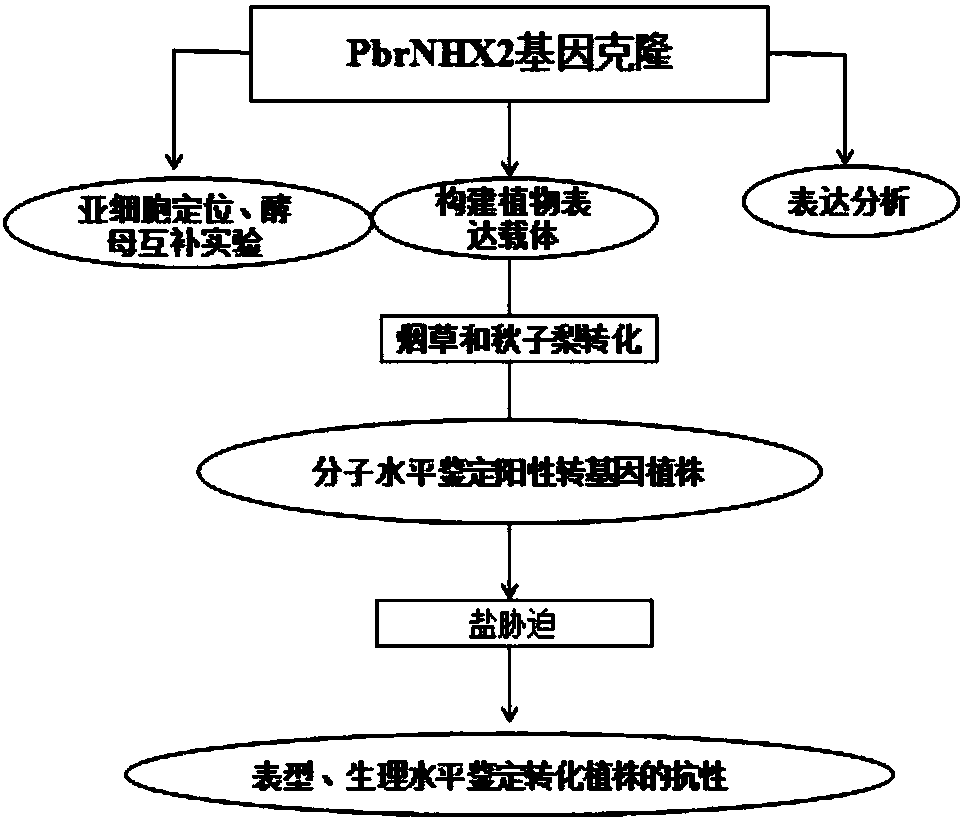
Sodium and hydrogen reverse transport protein PbrNHX2 in birch-leaf pears and application thereof in enhancement of salt resistance of plants
A technology of antiporter and Duli, applied in the field of genetic engineering, can solve the problems of complex genetic background, long childhood, self-incompatibility, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Cloning of full-length cDNA of PbNHX2 gene in Du pear
[0049] In the early stage, our research group used a high-efficiency yeast expression system to screen a sodium-hydrogen antiporter PbrNHX2 from Duli pear. According to the sequence of the PbrNHX2 gene and primer premier 5.0, primers were designed, and its full length was amplified from Duli pear by RT-PCR. long. The detailed steps are as follows: take 1 μg of Du pear RNA, treat it with 1U of DNase I at 37°C for 30 minutes, and immediately place it on ice, add 1 μl of 50mM EDTA, treat it at 65°C for 10 minutes, and immediately place it on ice. The synthesis of the first strand of cDNA was carried out according to the operation manual of the TOYOBO reverse transcription kit. The resulting first-strand cDNA was used for amplification of the PbrNHX2 gene. PCR was completed according to the following procedure: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 30 s, annealing at 58°C for 90 s, extension at...
Embodiment 2
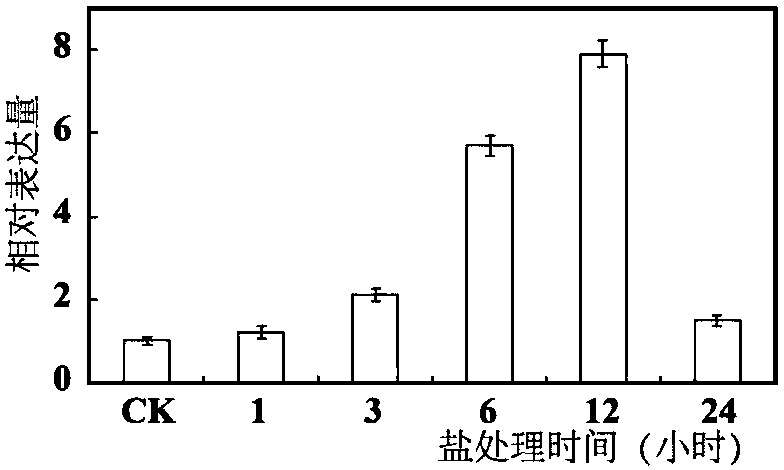
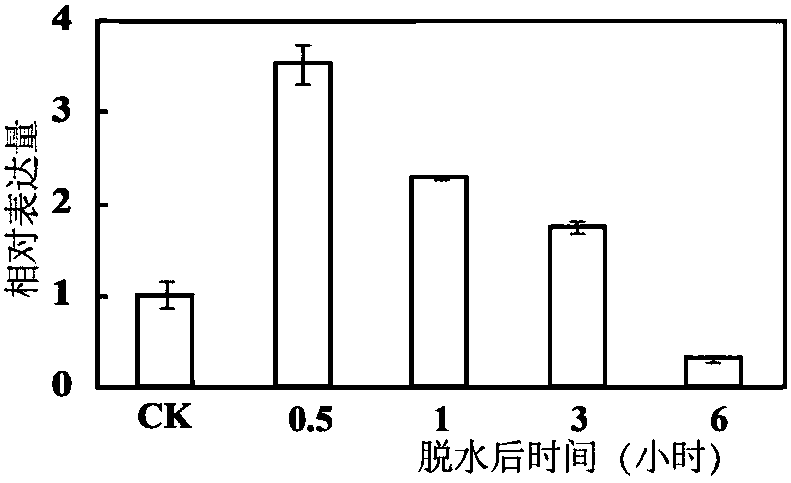
[0053] qRT-PCR analysis of PbrNHX2 gene and subcellular localization of PbrNHX2 gene under different stress conditions
[0054] In order to analyze the response pattern of PbrNHX2 gene to low temperature, high salt and dehydration in Duli pear, the expression pattern of PbrNHX2 gene was analyzed using Real-time PCR technology. The RNA was extracted by the CTAB method, and the synthesis of the first strand of DNA was carried out according to the operation manual of the TOYOBO reverse transcription kit. In the 20 μl reaction system, there are: 10 μl 2×Mix, 0.1 μl cDNA, 5 μl primers (using ubiqutin as internal reference primers (SEQ ID NO.5 and SEQ ID NO.6), the length is 208), 4.9 μl water. The program of quantitative PCR is shown in Table 1:
[0055] Table 1 Quantitative PCR program
[0056]
[0057] The results are shown in Figure 2. Fig. 2 is a schematic representation of expression of a PbNHX2 encoding gene of the present invention under salt, dehydration and low tempe...
Embodiment 3
[0059] Subcellular localization of the gene encoding PbrNHX2
[0060] According to the nucleotide sequence of the coding gene of PbrNHX2 and the pJIT166-GFP vector map, SalI and BamHI restriction sites were added before and after the gene sequence. The target gene extraction plasmid with correct sequencing results was used as a template, and amplified with primers (SEQ ID NO.7 and SEQ ID NO.8) with added restriction sites. The PCR program used was: 94°C pre-denaturation for 3 minutes; 94°C denaturation 30s, 58°C annealing for 1min, 72°C extension for 1min 30s, 35 cycles; 72°C extension for 10min. The stop codon TAG was removed at the 3' of the gene in order to allow the gene to be fused to GFP. After the PCR products were subjected to 1% agarose gel electrophoresis, the target band was recovered using a gel kit. The recovered and purified amplified fragment was cloned into pMD19-T vector, and transformed into Escherichia coli competent DH5α. The transformed bacterial liquid...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com