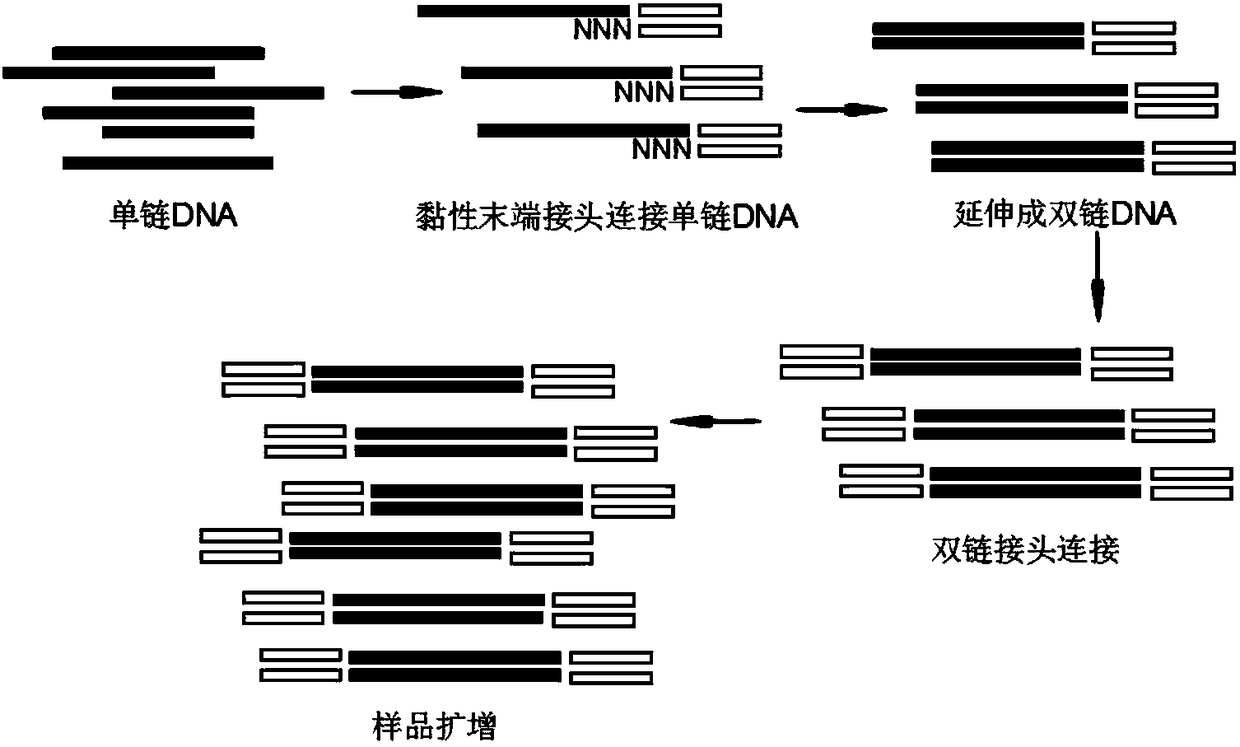
Novel DNA library construction method and application thereof
Another sticky-end technology, applied in the field of molecular biology, can solve the problems of loss of DNA information, inability to use DNA information effectively, and poor linking efficiency of adapters.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0202] 1. Test preparation
[0203] (1) Synthesis of cohesive end adapters:
[0204] The sticky end adapter is a double-stranded linker with a sticky end, one end of the sticky end adapter is a first sticky end, and the first sticky end includes at least a random sequence.
[0205] 1. As an example, use Primer1 and Primer2 to synthesize sticky end adapters,
[0206] Primer1: 5'-AGATCGGAAG-3'-bioten (SEQ ID NO.1);
[0207] Primer2: 5'-CTTCCGATCTNNNNNN-3' (SEQ ID NO.2); wherein, NNNNNN represents a random sequence, usually the length of the random sequence is 2-30nt, preferably 4-20nt, more preferably 5-10nt .
[0208] 2. Add the configuration reaction solution according to Table 1:
[0209] Table 1
[0210] Reagent
Volume (ul)
Primer1 (10μM)
1
Primer2 (10μM)
1
7
5M NaCl
1
total capacity
10
[0211] The TE buffer used in the reaction can also be replaced by water, Tris-Hcl (0.05mM, pH7.0-8.0) and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com