Nucleic acid amplification fluorescent quantitation method for detecting and integrating HIV-1 virus genomes and application thereof
A virus genome and HIV-1 technology, applied in the field of diagnosis, can solve the problems of rapid virus rebound and inaccuracy, and achieve the effects of high coverage, accurate quantitative results and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
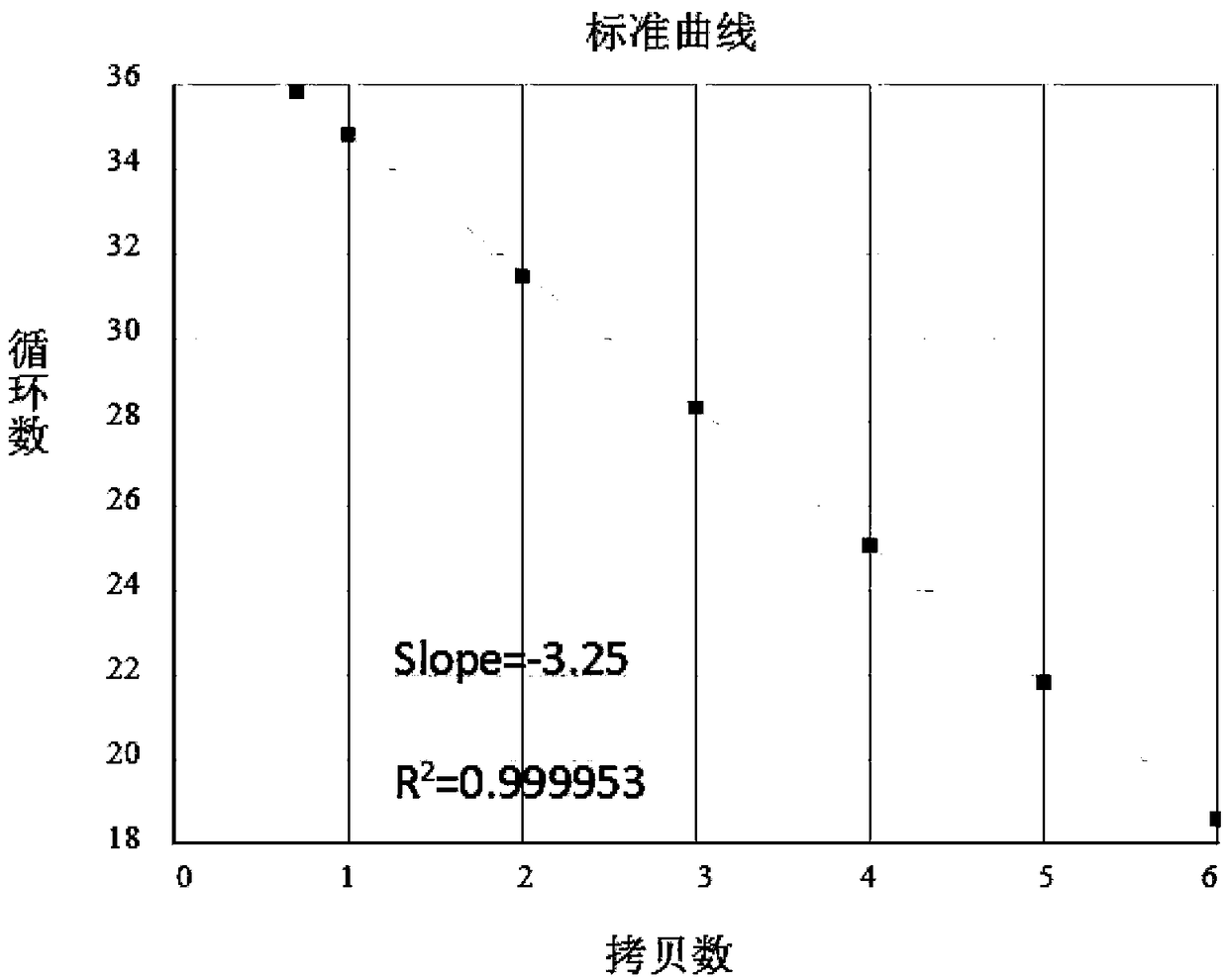
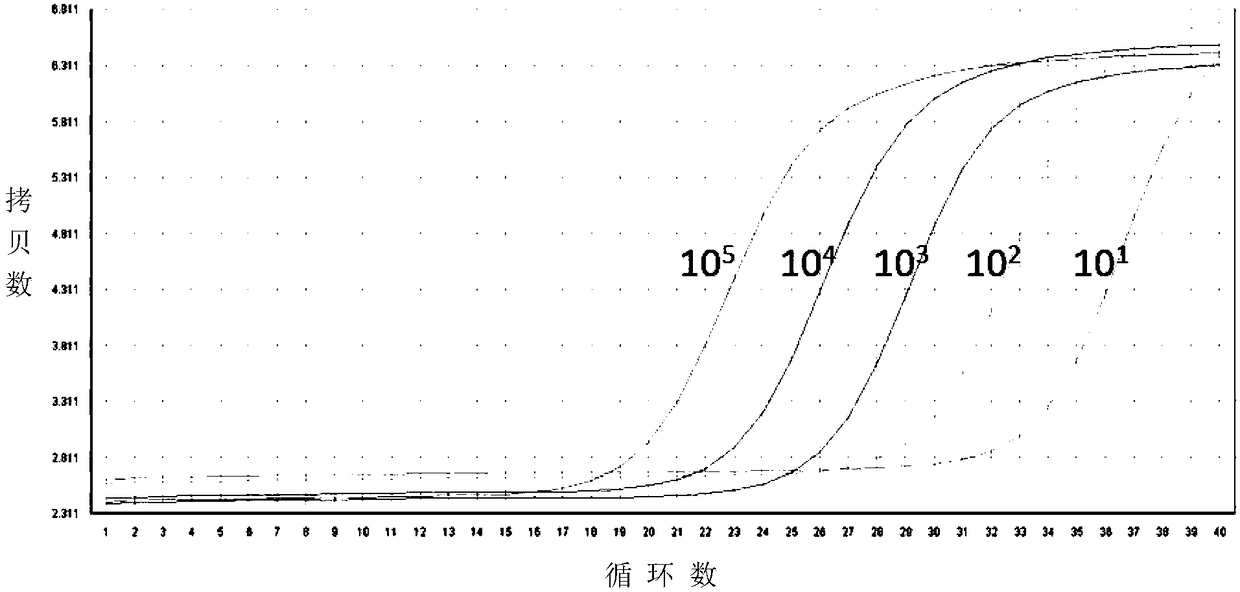
[0037] Example 1: Establishment of a nucleic acid amplification fluorescence quantitative method for detecting integrated HIV-1 virus genome
[0038] The nucleic acid amplification fluorescence quantitative method for detecting integrated HIV-1 virus genome of the present invention is divided into 4 steps: 1) Design and synthesize PCR amplification primers and TaqMan probes in the highly conserved Gag region of HIV virus sequences, and use gradient dilutions of known The plasmid DNA of copy number is template, establishes the method for real-time absolute quantitative HIV total DNA; 2) prepare the standard substance that is used to integrate HIV genome and adopt the method quantification of step 1); 4) Prepare the HIV integration standard and the test sample at the same time, with the test sample without Alu primer as the reference sample, through the same nested fluorescent quantitative PCR operation as step 3), and The standard curve is compared and the absolute copy number ...
Embodiment 2
[0079] Example 2: Verification of the effectiveness of nested fluorescent quantitative PCR in the detection of HIV integration provirus
[0080] 1. Experimental Design
[0081] In HIV-infected samples, various forms of viral DNA co-exist. In order to verify the specificity of the method, the experiment collected whole blood from healthy people and isolated peripheral blood lymphocytes (PBLs), respectively, with reverse transcriptase inhibitors. After two hours of treatment with Abacavir (150 μM) and integrase inhibitor Raltegravir (50 nM), the cells were infected at MOI=3 and incubated for 2 hours, washed twice with PBS, and placed in CO 2 The incubator continued to cultivate for 48 hours, and the cells were collected by centrifugation and washed. Then use the TIANamp Genomic DNAKit from QIAGEN to extract DNA, see the manual for details.
[0082] According to the steps and methods of Example 1, the quantification of the integrated HIV provirus was carried out, and the amplif...
Embodiment 3
[0085] Example 3: Quantitative detection of HIV-integrated provirus in blood samples of HIV-1 infected persons
[0086] 1. Processing of samples (conducted in BSL-2+ level laboratory)
[0087] Blood samples from 14 HIV-1 infected patients were collected after the informed consent was signed by the hospital ethics committee.
[0088] Separation of peripheral blood mononuclear cells (PBMC): 10ml of EDTA anticoagulant venous blood was drawn from 14 HIV-infected patients, and the specimens were collected in accordance with the "National Technical Specifications for AIDS Testing", and separated by lymphocytes within 4 hours after blood collection PBMCs were obtained by separating them with Ficoll, and DNA was extracted using TIANampGenomic DNA Kit from QIAGEN. For details, please refer to the instruction manual.
[0089] 2. Detection of HIV integrated provirus in blood samples by nested fluorescent quantitative PCR
[0090] According to the method of step 1) in the technical solu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com