SNP molecular marker for identifying and controlling formation of low-temperature yellowing property of Chinese cabbage leaves and application thereof
A DNA molecule, low temperature technology, applied in the biological field, to achieve the effect of high-efficiency assisted breeding methods and technologies, high specificity and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1, the development of SNP molecular marker
[0063] Resequencing of the low-temperature-resistant Chinese cabbage material (ST64) and the low-temperature-resistant Chinese cabbage material (ST336), after data analysis, it was found that there was a SNP site at the 13595848th nucleotide of the A05 chromosome (the 300th in the sequence listing sequence 4 nucleotide), which is a T / G polymorphism, and the SNP site is named A05-13595848T / G site.
[0064] For the A05-13595848T / G locus, based on the principle of competitive allele-specific PCR, the primers were designed as follows:
[0065] Upstream primer A05-13595848T / G-FF (sequence 1 of the sequence listing): 5'- GAAGGTGACCAAGT TCATGCTGGAGGGGTTGTTGGGACGg-3';
[0066] Upstream primer A05-13595848T / G-FV (sequence 2 of the sequence listing): 5'- GAAGGTCGGAGT CAACGGATTGGGAGGGGTTGTTGGGACGt-3':
[0067] Downstream primer A05-13595848T / G-R (sequence 3 in the sequence listing): 5'-CACTCATCGCCGTCCACGCGTA-3'.
[006...
Embodiment 2
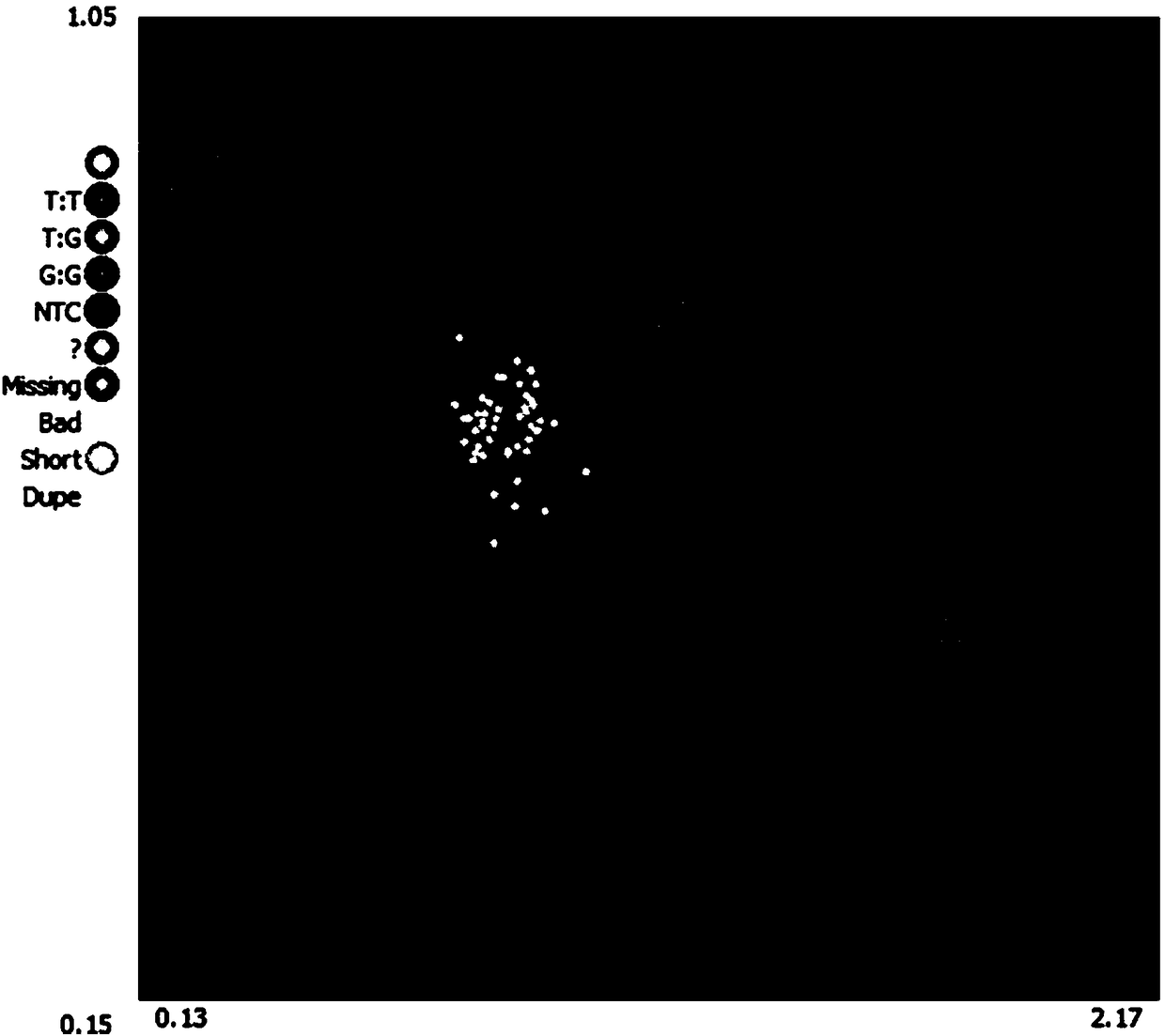
[0069] Example 2. Application of A05-13595848T / G Loci in Identifying Yellowing and Non-yellowing Materials of Chinese Cabbage After Low Temperature Induction
[0070] 1. DNA extraction
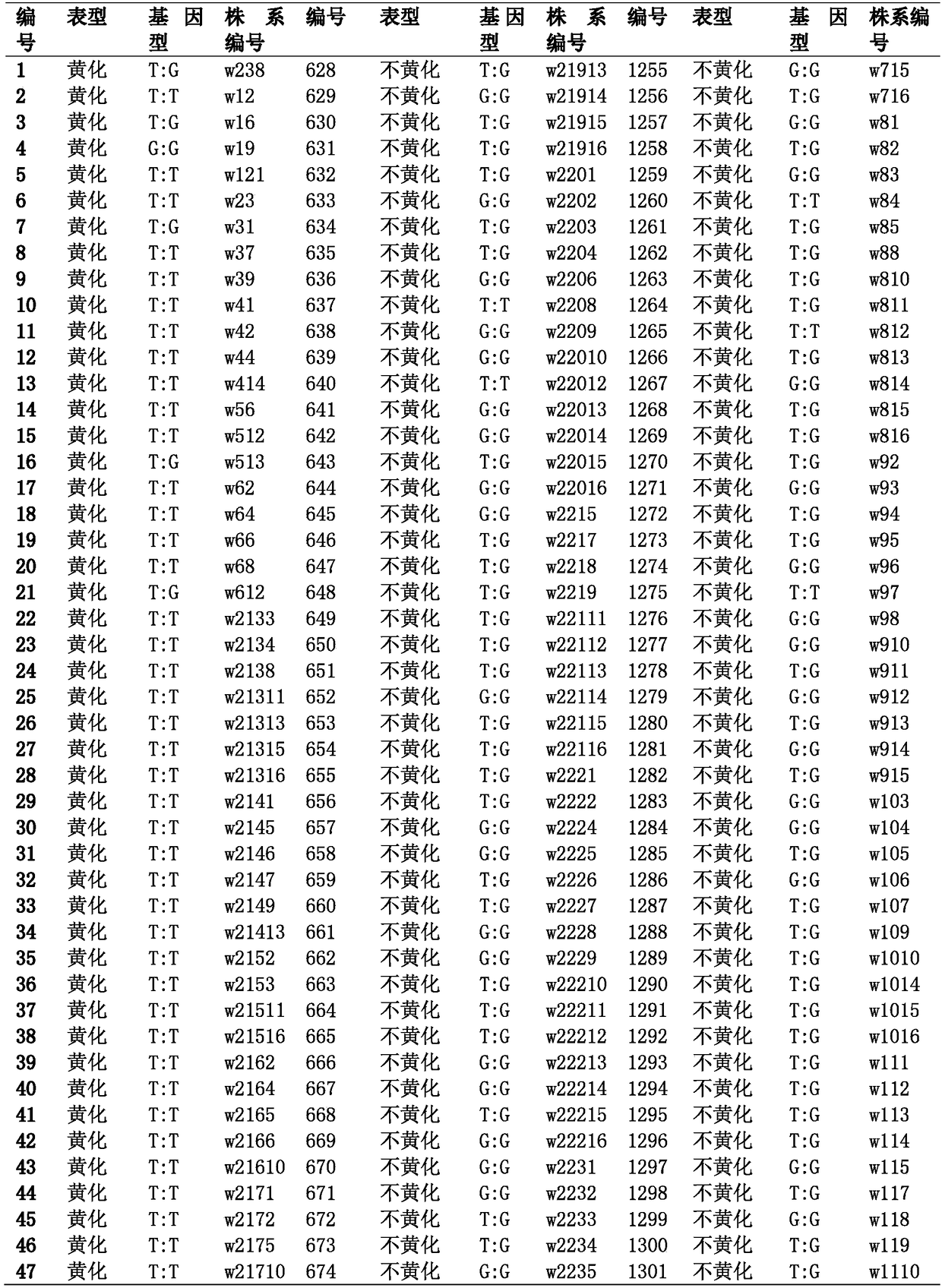
[0071] Genomic DNA of the 1881 cabbage materials to be tested in Table 1 were extracted by conventional CTAB method.
[0072] 1881 materials are the hybrid offspring F of parents ST64 and ST336 2 For each individual plant, the specific method is as follows: cross the low-temperature non-yellowing parent material ST64 and the low-temperature yellowing parent material ST336 to obtain F 1 ;F 1 Continue to inbred to get F 2 group.
[0073] The quality of the extracted DNA was tested by agarose electrophoresis and Nanodrop2100 respectively, and it was found that the extracted genomic DNA met the relevant quality requirements, that is, agarose electrophoresis showed a single DNA band without obvious dispersion; Nanodrop2100 detected A260 / 280 between 1.8-2.0 between (DNA samples without protein ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com