SNP molecular marker for cold-resistance identification of apple rootstocks, primers and application of SNP molecular marker
A technology of molecular markers and cold resistance, applied in the field of crop genetics and breeding, can solve the problems of long cycle, achieve accurate identification, reduce time and economic cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The SNP molecular marker of apple rootstock cold resistance identification of the present invention adopts the following method to obtain:
[0035] 1. Reagents and instruments used
[0036] Genomic DNA (gDNA) of individuals in the test population was extracted using the Plant Genomic DNA Kit (TIANGEN) kit, which was purchased from Tiangen Biochemical Technology (Beijing) Co., Ltd.; restriction enzymes for enzyme digestion EcoRI Purchased from Sigma-Aldrich Company in the United States; reagents required for post-processing of enzyme-digested samples: adapters, ATP, ligase, blunt-end enzyme, etc. were purchased from Illumina Company in the United States; The IlluminaHiseq 2500 sequencing platform (USA) was used.
[0037] 2. SNP molecular marker acquisition method
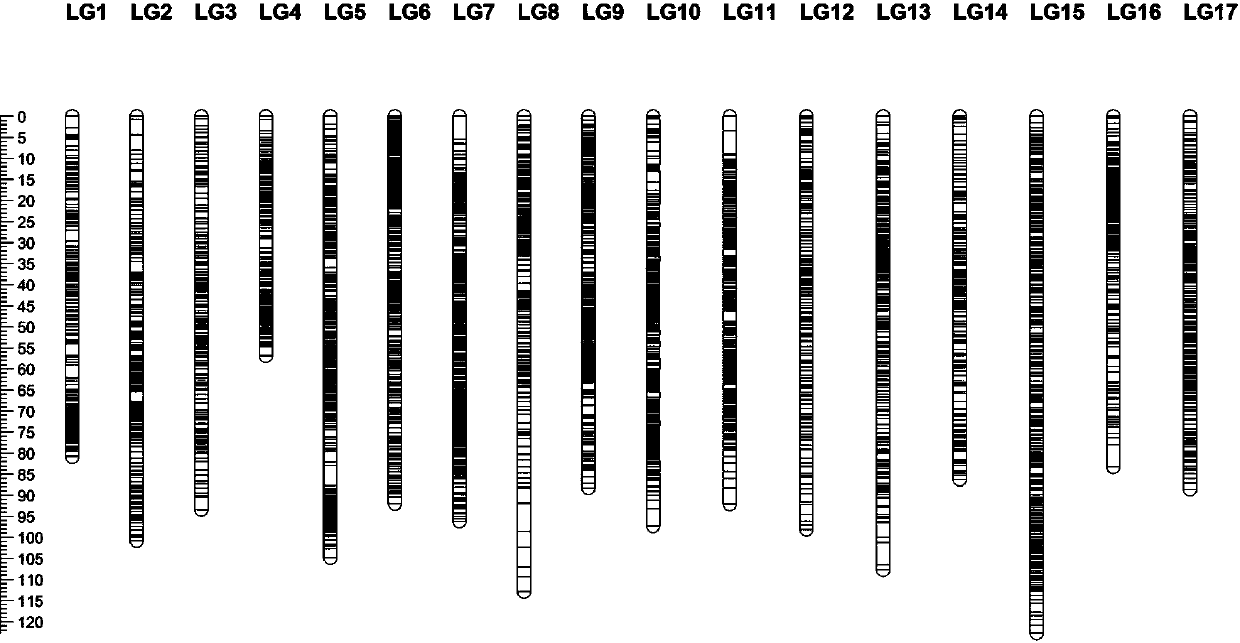
[0038] (1) "60-160×M9" genetic linkage map construction
[0039] The hybrid offspring of apple cold-resistant rootstock "60-160" (low temperature semi-lethal temperature -37.42C°) x non-cold-resistant root...
Embodiment 2
[0058] In order to efficiently, quickly and accurately detect the SNP molecular marker of the present invention, it can be made into a SNP gene chip. The above-mentioned PCR primers for amplifying the SNP molecular markers are fixed on the SNP gene chip, and the gene chip is manufactured by Affymetrix Company of the United States. The genomic DNA of the test population was extracted, KASP reaction was performed on the SNP gene chip, the SNP molecular marker was amplified, and the cold resistance performance of the test population was quickly identified through the genotype of the SNP molecular marker. The specific operation is as follows:
[0059] Collect 182 "60-160" × "M9" mapping population strains, use the Plant Genomic DNA Extraction Kit (PlantGenomic DNA Kit, Beijing Tiangen Company) to extract the genomic DNA of the test population, and perform KASP on the SNP gene chip produced reaction. A total of 1 microliter of KASP amplification reaction system, including 20 nano...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com