Soluble microneedle patch and preparation method thereof
A probe and chip technology, applied in the field of soluble microneedle patches and its preparation, can solve the problems of poor detection ability of mixed genotypes, HBsAg antigenic changes, and difficulty in detecting multi-gene locus mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Embodiment 1, the preparation of gene chip
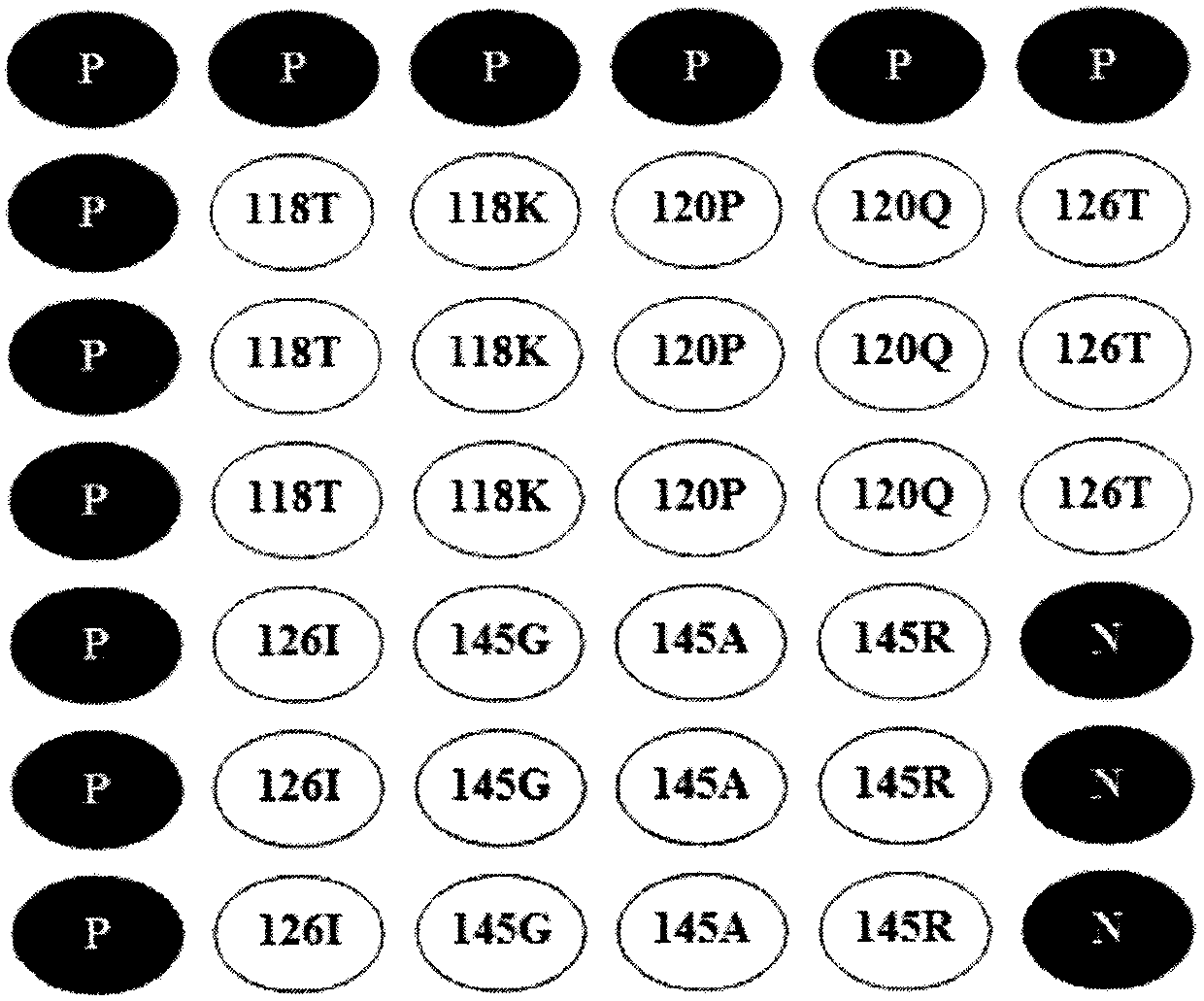
[0091] According to the characteristics of the prevalence of hepatitis B virus (HBV) sequences in my country, the oligonucleotide probes used to detect 4 HBsAg mutation sites are designed, as shown in Table 1 (the probes in Table 1 are all carried out at the 3' end. NH 2 modified).
[0092] The HBsAg wild type is shown in sequence 14 of the sequence listing. The four HBsAg mutation sites are: (1) S118: amino acid 118 of HBsAg; (2) S120: amino acid 120 of HBsAg; (3) S126: amino acid 126 of HBsAg; (4) S145: amino acid 145 of HBsAg bit amino acid.
[0093] Table 1 Probe information
[0094] probe name
Probe sequence (5'-3')
118T19
CTACCAGCACGGGACMATGTTTTTTTTTTTT (sequence 1)
31
118K19
CTACCAGCAAGGGACMATGTTTTTTTTTTTT (sequence 2)
31
120P17
CAMGGGACCATGCAAAATTTTTTTTTTTT (sequence 3)
29
120Q17
CAMGGGACAATGCAAAATTTTTTTTTTTT (sequence 4)
29
126...
Embodiment 2
[0104] Embodiment 2, establishment of detection method
[0105] 1. Take the serum to be tested, and use the virus DNA out kit to extract the virus DNA according to the instructions.
[0106] 2. Using the viral DNA obtained in step 1 as a template, carry out a nested PCR reaction to obtain a PCR amplification product (about 1200bp);
[0107] Specific steps are as follows:
[0108] (1) The first round of PCR, reaction system: template (viral DNA) 5 μl, PCR reaction solution 12 μl, upstream and downstream primers 2.5 μl each, ddH 2 O 2.5 μl, Taq enzyme 0.5 μl; PCR reaction program: pre-denaturation at 94°C for 3 minutes, denaturation at 94°C for 35 s, annealing at 59°C for 35 s, extension at 72°C for 70 s, 2 cycles; denaturation at 94°C for 35 s, annealing at 57°C for 35 s, and 72°C Extension 70s, 2 cycles; denaturation at 94°C for 35s, annealing at 55°C for 35s, extension at 72°C for 70s, 2 cycles; denaturation at 94°C for 35s, annealing at 53°C for 35s, extension at 72°C for ...
Embodiment 3
[0126] Embodiment 3, clinical sample detection
[0127] Research objects: 40 patients with chronic hepatitis B, 24 males and 16 females, in line with the diagnosis of chronic hepatitis B in the Chinese Medical Association Hepatology Branch, Chinese Medical Association Infectious Diseases Branch: Guidelines for the Prevention and Treatment of Chronic Hepatitis B (2015 Edition) Criteria, excluding autoimmune diseases and other liver diseases.
[0128] Serum samples were taken from the research subjects and tested according to the method in Example 2.
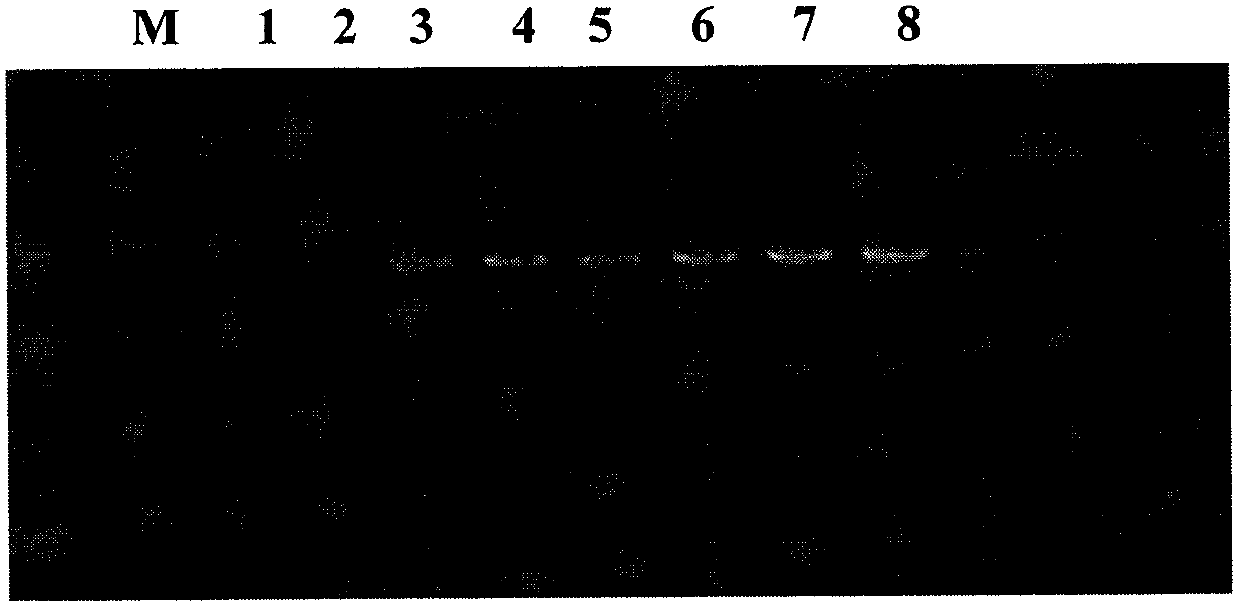
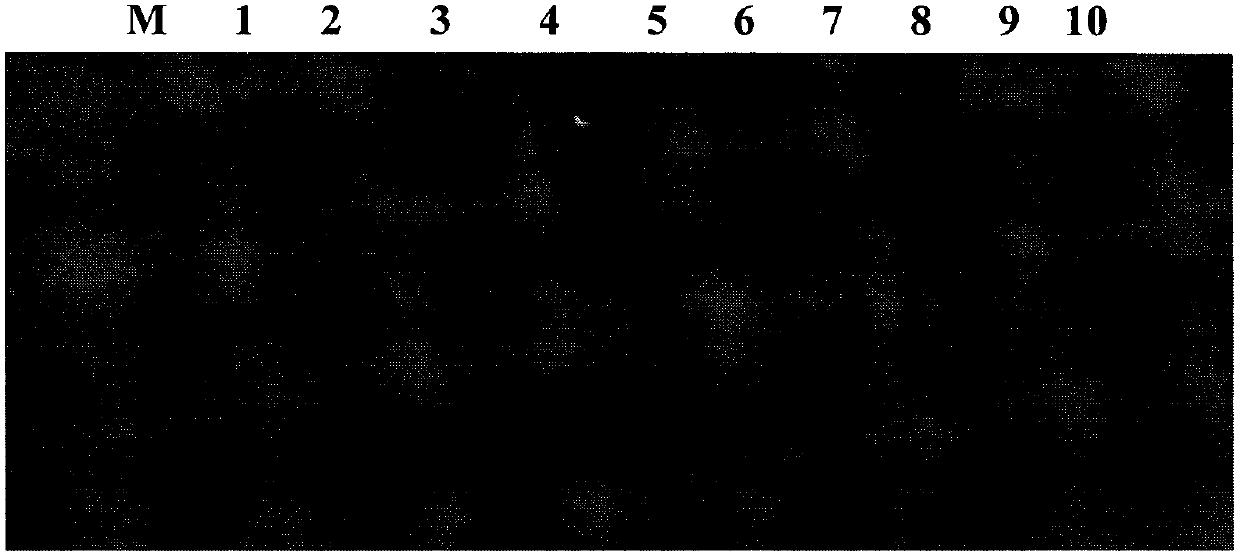
[0129] The electrophoresis diagram of the PCR amplification product obtained in step 2 of some samples is shown in figure 2 , the size of the amplified product is about 1200bp, which is consistent with the expectation; the electropherogram of the PCR amplified product obtained after step 6 is shown in image 3 , the size of the amplified products is about 300bp, which is consistent with the expectation.
[0130] A total of 40 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com