DNA Transmethylase defective CHO (Chinese hamster ovary) cell line and preparation method and application thereof
A methyltransferase and cell line technology, applied in the field of genetic engineering, can solve the problems of unstable expression, inability to meet vaccine requirements, and low expression level.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Knockout of CHO cell Dnmt3a gene
[0030] 1.1 Determine the target site for the candidate gene
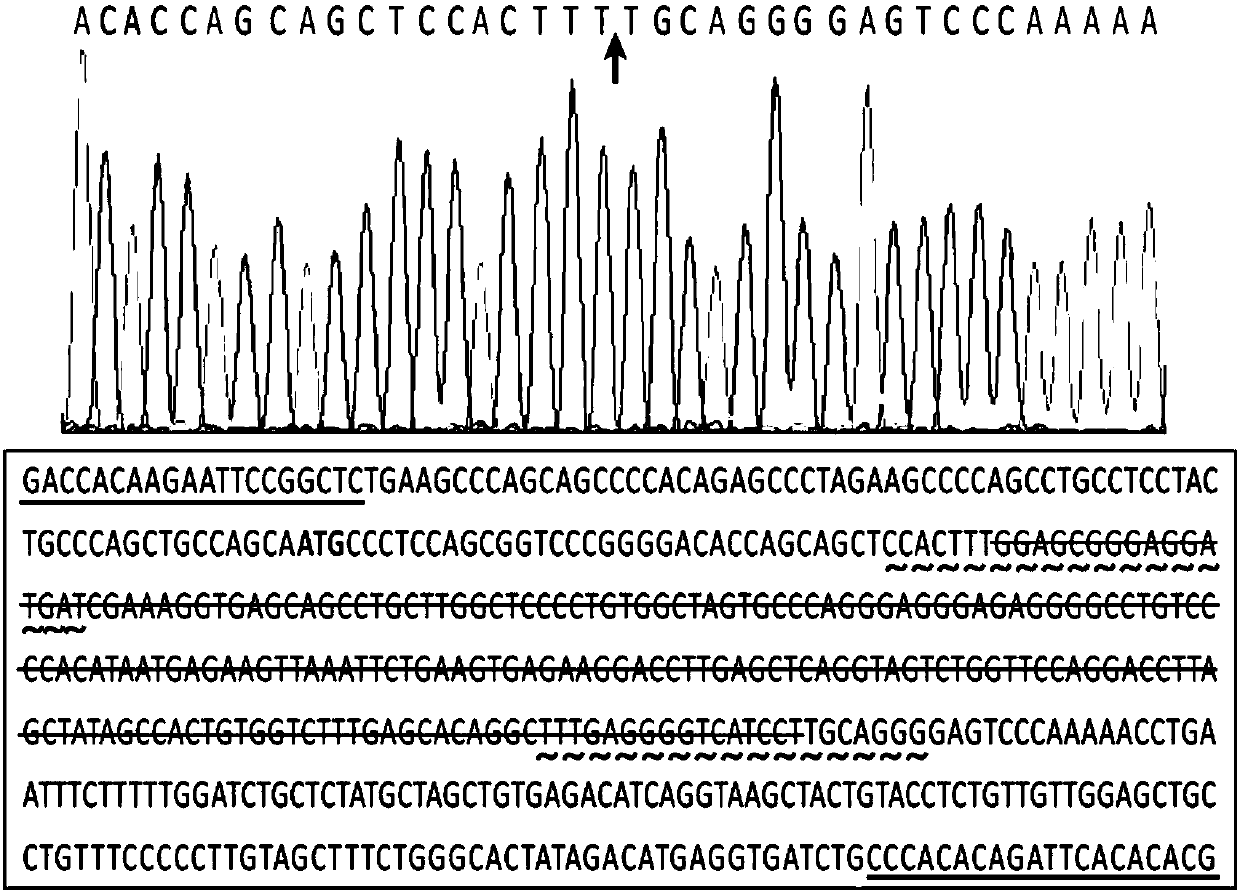
[0031]Primers (D3a-Ex1seq-L:5′-GACCACAAGAATTCCGGCTC-3′, D3a-Ex1seq-R: 5'-CGTGTGTGAATCTGTGTGGG-3') PCR amplification of the corresponding gene fragment was carried out, and the accuracy of the sequence was verified by cloning and sequencing of the PCR amplified fragment. The chimeric single-stranded guide RNA targeting site of the Dnmt3a gene assisted by the online tool (http: / / crispr.mit.edu / ) is as follows:
[0032] D3a-Ex1-98rev: 5'-ATCATCCTCCCGCTCCAAAGTGG-3';
[0033] D3a-Ex1-308fw:5′-TTTGAGGGGTCATCCTTGCAGGG3′
[0034] Two pairs of primers were designed according to the target sequence to construct the targeting sgRNA vector expressing CRISPR / Cas9.
[0035] 1.2 Construction of sgRNA vector
[0036] Use Bbs I restriction endonuclease to linearize the px330 vector plasmid and recover the linear fragment. The sgRNA oligonucleotide single strand anneals to for...
Embodiment 2
[0039] Example 2 Verification of biological characteristics of Dnmt3a-deficient CHO cells
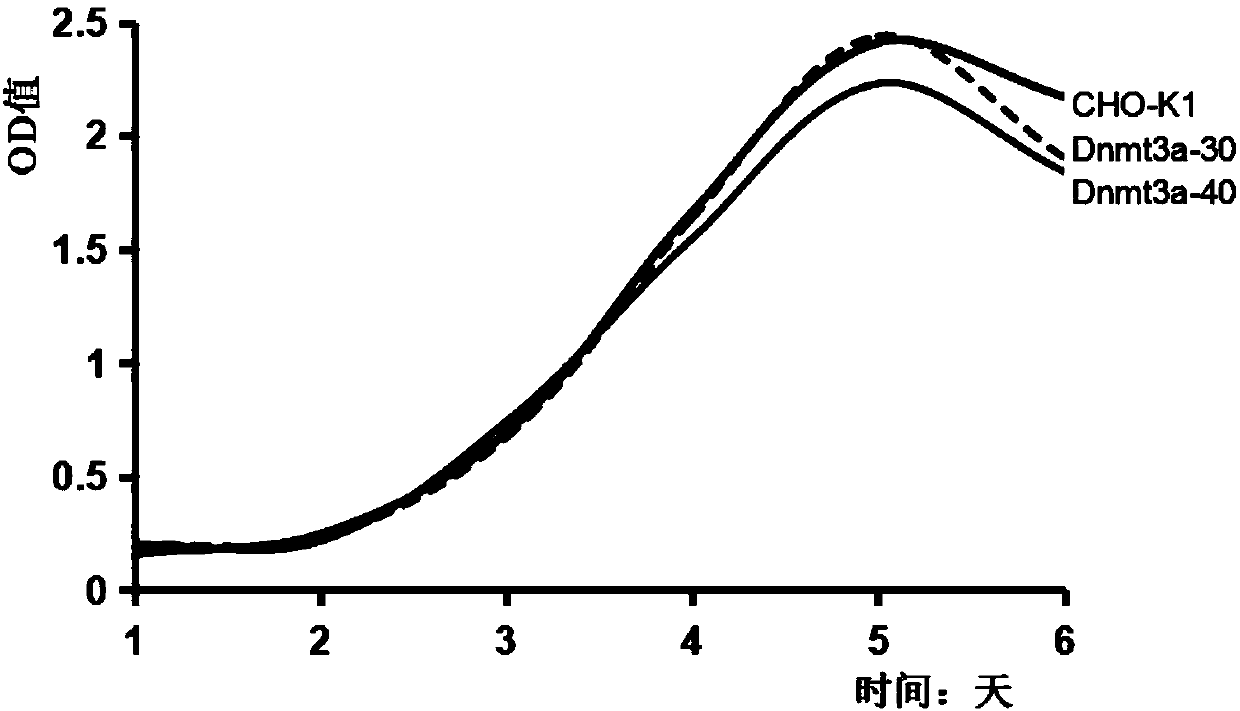
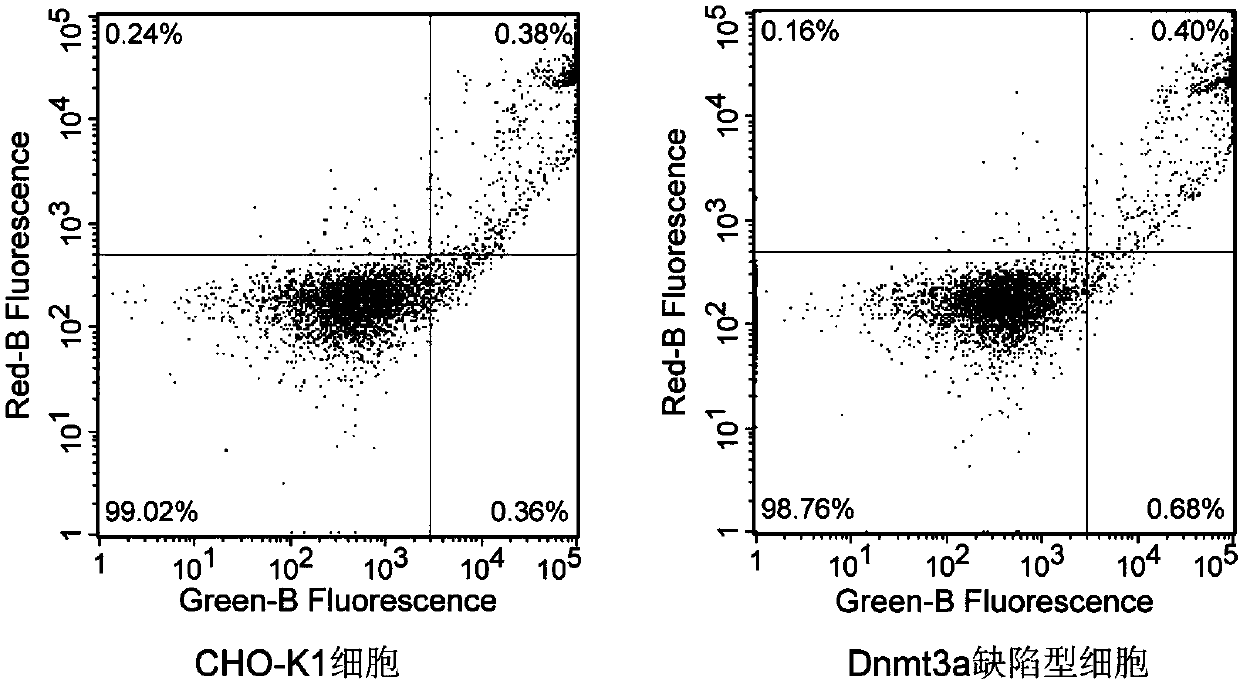
[0040] Using wild-type CHO-K1 cells as a control, the cell growth characteristics of the obtained Dnmt3a-deficient CHO cell clones were verified, including observation of cell morphology and growth status, detection of cell proliferation by CCK-8 method, flow cytometry (FCM) Cell apoptosis was detected to verify whether the Dnmt3a-deficient CHO cell line could undergo normal growth and passage. CCK-8 kit (Cell Counting Kit-8 kit, Beyotime Biological Company) detects cell proliferation (results see figure 2 ) and the results of cell apoptosis detected by flow cytometry (results in image 3 ) suggest that the obtained Dnmt3a-deficient CHO cell line can grow and pass on normally, and the biological characteristics such as cell growth state, morphology, cell proliferation, and cell apoptosis have no significant difference from normal CHO cells.
Embodiment 3
[0041] Example 3 Construction of recombinant expression vectors driven by different promoters
[0042] The present invention uses the eukaryotic expression vector pIRESneo2 (Clontech company) as the parent carrier to construct the eukaryotic expression vector pWTY-02 (see Figure 4 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com