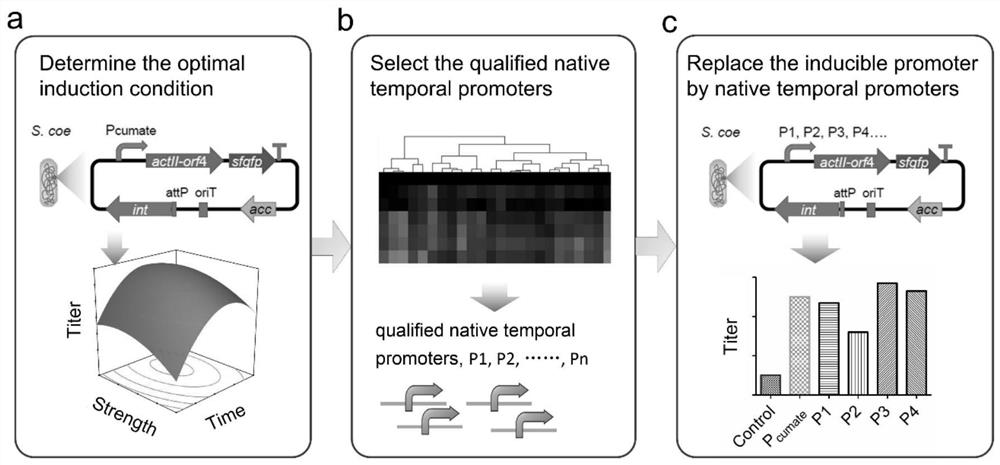
A method for increasing the yield of streptomyces secondary metabolites
A secondary metabolite, Streptomyces coelicolor technology, applied in the field of genetic engineering and fermentation engineering, can solve the problems of increasing cost and difficulty in product purification, and achieve the effect of increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] The construction of a kind of genetically engineered bacteria of embodiment 1
[0016] Use primers P1-F / P1-R and plasmid pIJ8660::BsaI-sfgfp as a template to amplify the plasmid backbone; use the Streptomyces coelicolor M145 genome as a template and primers actII4-F / actII4-R to amplify actII-orf4 Gene, and the above-mentioned linear plasmid backbone using the Gibson assembly method to obtain the plasmid pActII-sfgfp. Using the Streptomyces coelicolor M145 genome as a template, the corresponding primers were used to amplify the physiological promoter. After digestion with BglII and EcoRV, it was digested with the corresponding enzymes and ligated with the plasmid pActII-sfgfp to obtain the plasmid pPn-actII-sfgfp in which the expression of the actII-orf4 gene is controlled by a physiological promoter. Using the plasmid pGCymRP21 as a template, use the primers cumate-F / cumate-R to amplify the cumate-inducible promoter, digest it with BglII and EcoRV, and then connect it ...
Embodiment 2
[0018] The construction of a kind of genetically engineered bacteria of embodiment 2
[0019] Using the plasmid pIJ8660::BsaI-sfgfp as a template, the plasmid backbone was amplified using primers P1-F / P1-R. Using Streptomyces chamobilis genome as a template and primers otcR-F / otcR-R, the otcR gene was amplified, and combined with the above-mentioned linear plasmid backbone using the Gibson assembly method to obtain the plasmid pOtcR-sfgfp. Using Streptomyces coelicolor as a template, using the corresponding primers to amplify the appropriate physiological promoters, the plasmid pOtcR-sfgfp and these promoters were digested with BglII and EcoRV and ligated to obtain the plasmid pPn that uses the physiological promoter to control the otcR gene -otcR-sfgfp. Similarly, the digested kasO* and cumate promoter fragments were connected into the digested linear plasmid pOtcR-sfgfp to obtain plasmids pK-otcR-sfgfp and pCumate- otcR-sfgfp. Plasmids pPn-otcR-sfgfp and pCumate-otcR-sfgf...
Embodiment 3
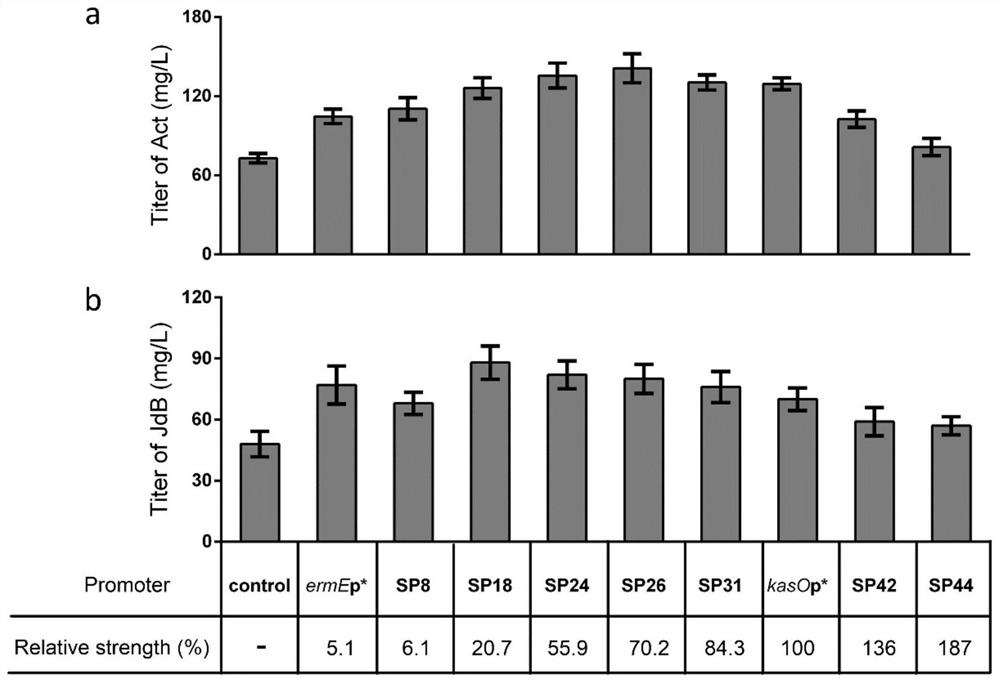
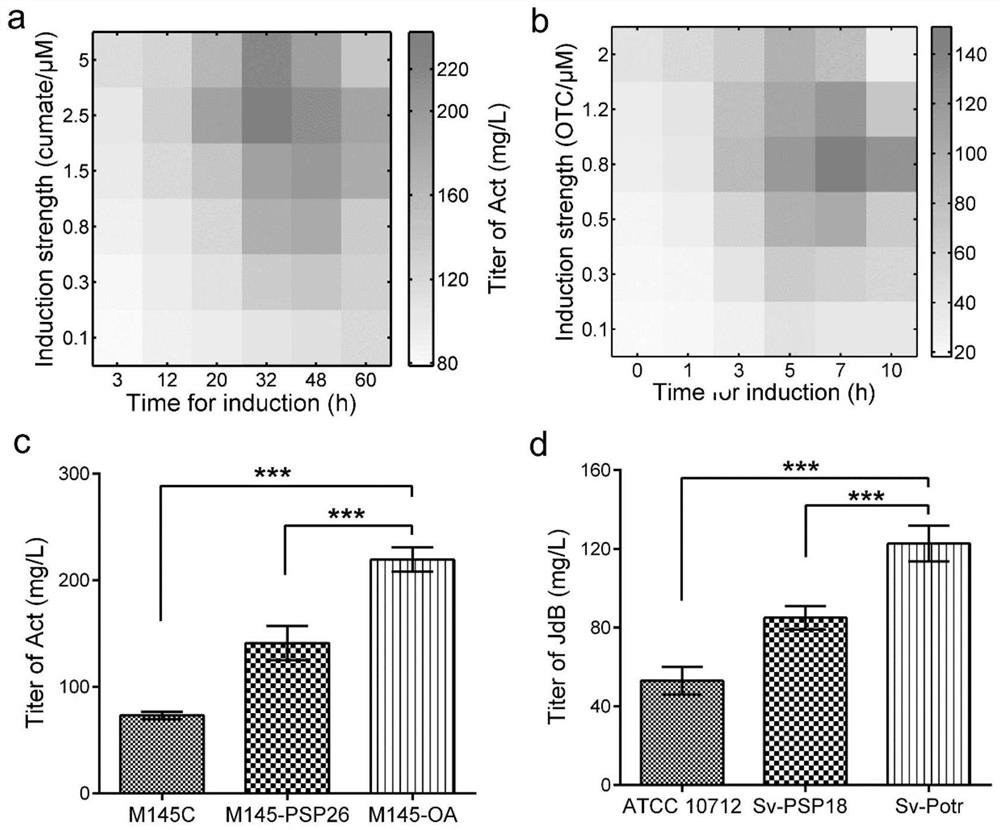
[0029] Example 3 Production of secondary metabolites actin and jodomycin B by fermentation and analysis of results
[0030] Utilize the genetically engineered bacteria M145-Pc prepared in Example 1, M145-OA, M145-Pn to ferment and produce the secondary metabolite actinhodine, the specific process is: Streptomyces coelicolor engineering strains are placed on MS solid plate (2% Mannitol, 2% soybean flour, 2% agar) were cultured at 28°C. After collecting the spores, press 4×10 8 The concentration of spores / 100ml was inoculated into 50mL SMM medium (81.9ml PEG6000 (6.1%, w / v), 2.5ml magnesium sulfate (24g / L), 10ml TES buffer (0.25M, pH7.2) , 2ml glucose (50%, w / v), 0.1ml trace elements (zinc sulfate, ferrous sulfate, manganese chloride, calcium chloride, sodium chloride each 0.1g / L), 1ml casein hydrolyzate (20% , w / v), 25ml glycine (20%, w / v)) was fermented in a 250mL shake flask at 28°C, 250rpm. Collect the bacteria, centrifuge at 10,000×g at 4°C for 3 minutes, collect the sup...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com