Cell strain capable of reducing replicability adenovirus production and construction method and application thereof
A construction method and cell line technology, applied in the field of cell lines and construction to reduce the production of replicable adenovirus, can solve the problem of reduction and achieve the effect of reducing off-target effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Construction of Crispr / Cas9n system for HEK293 cell E1 gene ITR and E1 Promoter
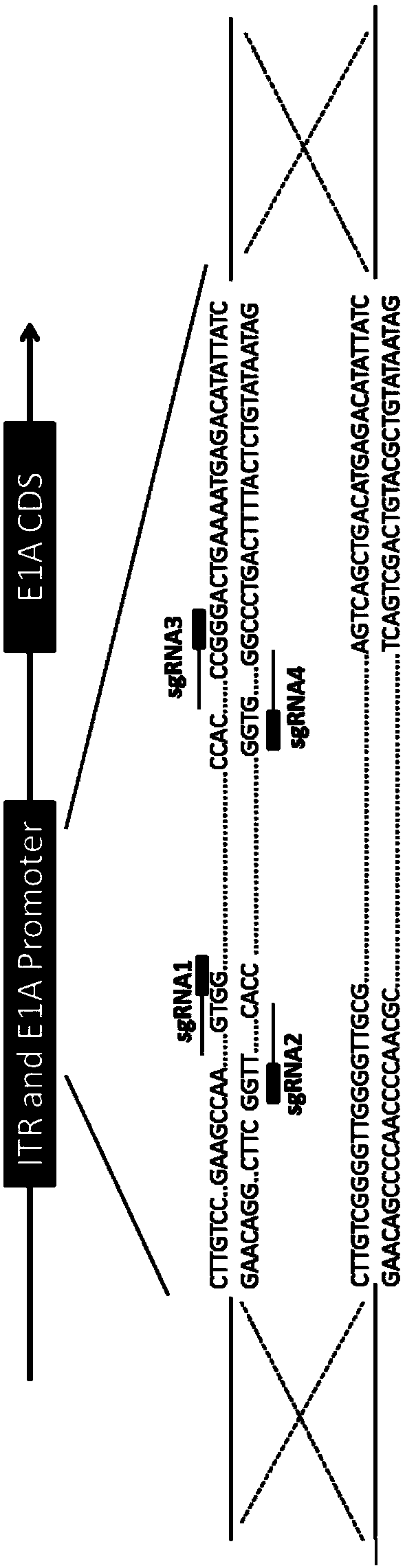
[0054] First, according to the HEK293 genome sequence in NCBI, select the E1 gene ITR E1 Promoter as the target site to design sgRNA, and the target site sequence is as follows figure 1 As shown, the sgRNA sequence is shown in Table 1 above.
[0055] Secondly, the construction of PX462.V2.0-sgRNA containing specific sgRNA sequence:
[0056] (1) Design and synthesize sgRNA that recognizes E1 gene ITR and E1 Promoter;
[0057] (2) The synthesized sgRNA oligonucleotides are annealed in vitro;
[0058] (3) Digest PX462.V2.0 through the Bsa I site and connect with sgRNA, named PX462.V2.0-sgRNA.
[0059] Finally, a homology repair template plasmid was designed according to the target site sequence of the sgRNA.
Embodiment 2
[0061] cell transfection
[0062] 4 × 10 in a six-well plate 5 Inoculate HEK293 cells at a density of 70%-90% (about 18-20h) per well, and start transfection when they grow to a fusion rate of 70%-90% (about 18-20h). The concentration of PX462.V2.0-sgRNA1, PX462.V2.0-sgRNA2, PX462.V2.0-sgRNA3, PX462.V2.0-sgRNA4 is 20ng / μL, and the PGK Promoter repair template plasmid is 25ng / μL), Take 20 μL and transfect cells with 5 μL of Lipo2000 transfection reagent, add SCR7 non-homologous recombination inhibitor (final concentration: 0.01 mM) after 12 hours, and add Puromycin (final concentration: 3 μg / mL) after 36 hours for screening.
Embodiment 3
[0064] screening verification
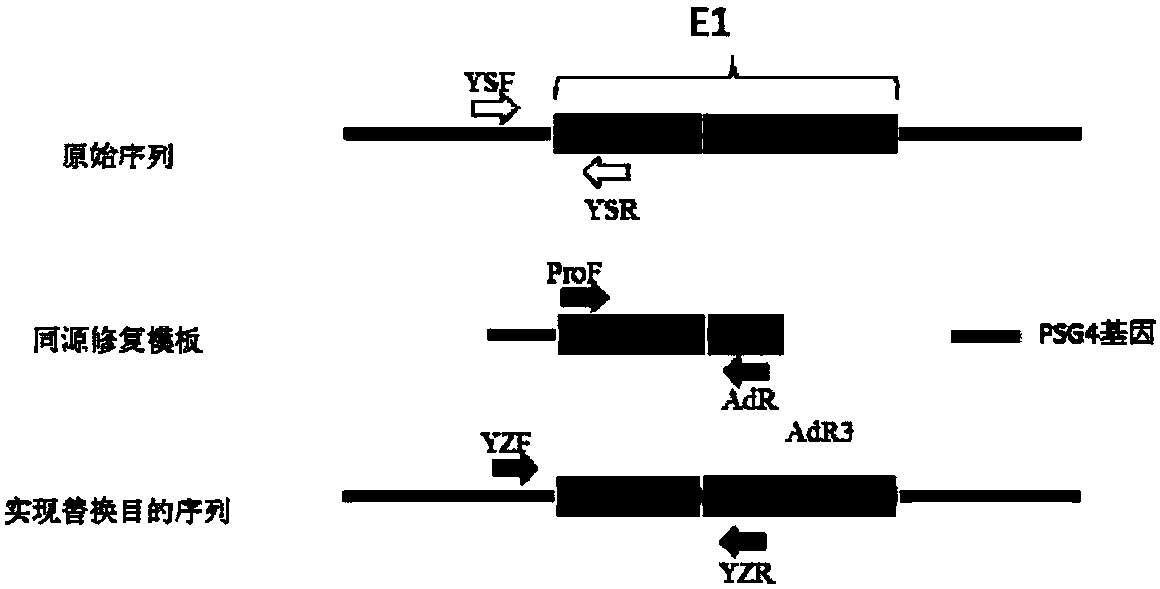
[0065] use image 3 The method shown verifies whether the E1 gene in HEK293.CS cells has been completely replaced. The method is: extract the genome of the transformed cells, and if the ProF / AdR primer pair can amplify bands, it proves that the original sequence is replaced by PGK Promoter, such as YSF The / R primer pair can amplify the fragment, which proves that the original sequence still exists in the replaced cells, and the replacement is not complete. Only when the ProF / AdR primer pair can amplify the band but the YSF / R primer pair cannot amplify the band. It proved that ITR and E1A Promoter were completely replaced by PGK Promoter in HEK293 cells. If the sequencing result of the fragment amplified by the YZF / R primer pair is a pure PGK Promoter sequence, it further proves that the original sequence has been completely replaced.
[0066] The verification primers are shown in Table 2, and the sequencing results are shown in Figure 4 It ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com