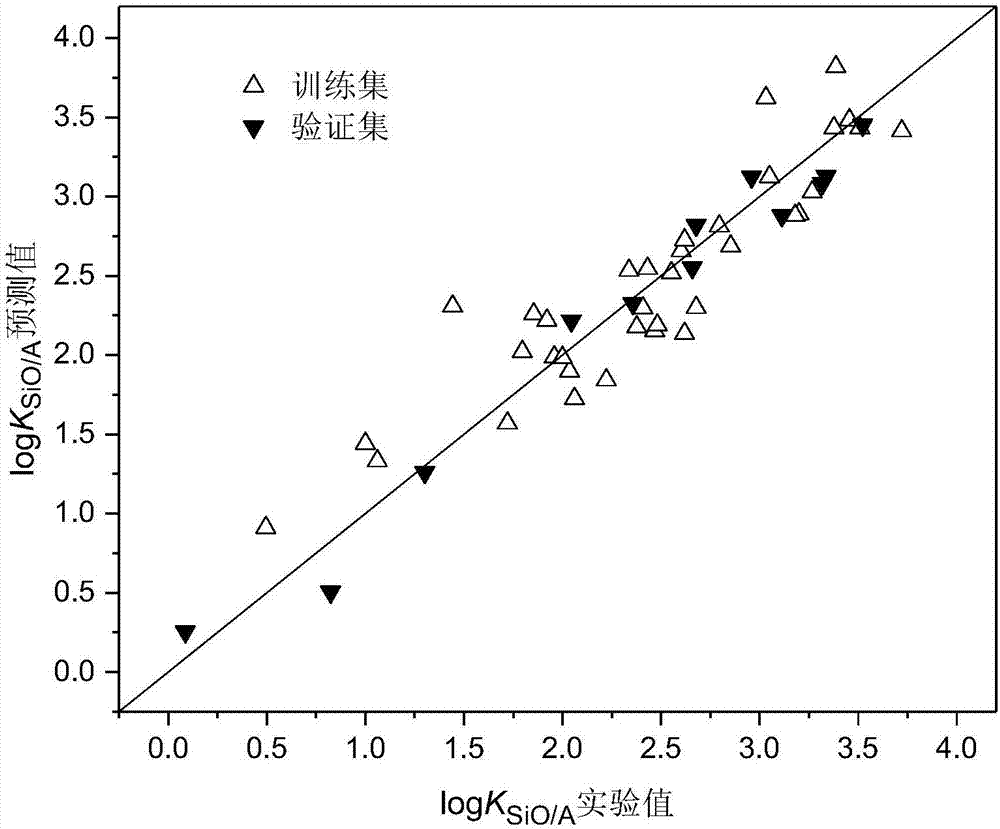
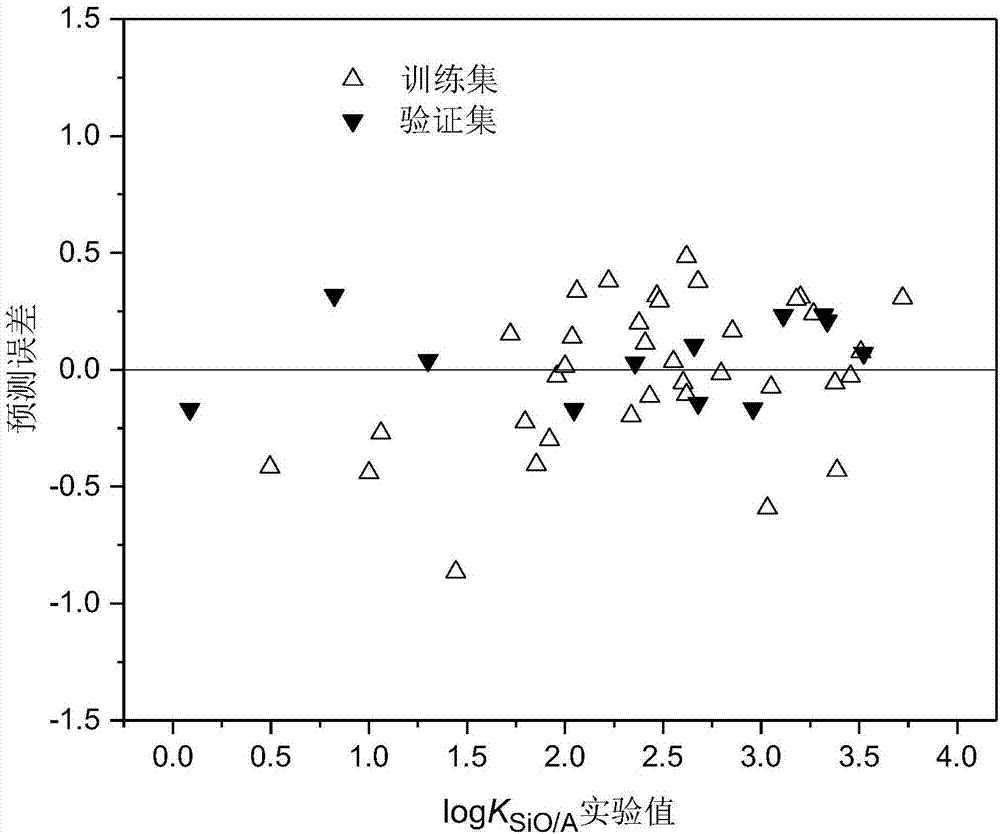
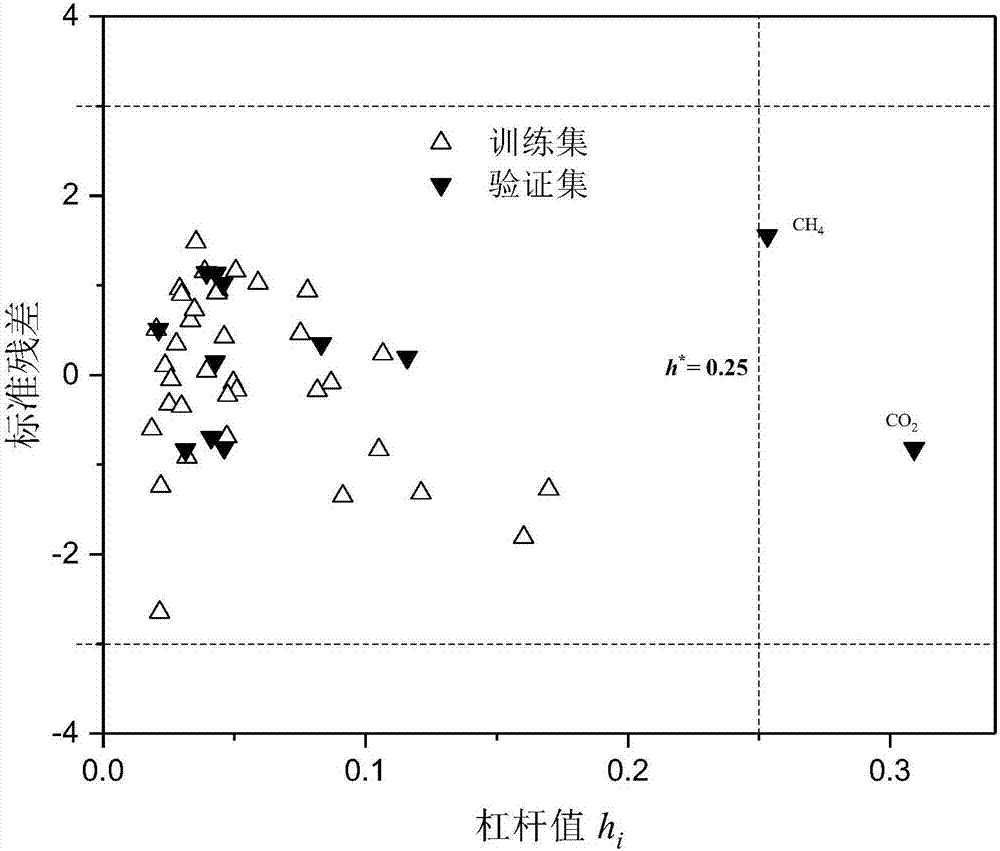
Method for constructing quantitative structure-activity relationship model to predict silicone oil-air partition coefficient of hydrophobic compound
A technique for hydrophobic compounds and quantitative structure-activity relationships, applied in chemical data mining, chemical statistics, chemical machine learning, etc. It is easy to understand and apply, save manpower, and low cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Dimethyl sulfide: First, check the molecular structure information of dimethyl sulfide on the organic small molecule biological activity database (PubChem), and then use the B3LYP / 6-311G** method in the quantum chemistry software Gaussian to calculate α, E LUMO -E HOMO These 2 descriptors. Its h is calculated by Williams graph method i The value is 0.024-3, indicating that this compound is within the application domain of the QSAR model constructed in the specific embodiment of the present invention.
[0056] Substituting into the constructed QSAR model, the logK of dimethyl sulfide SiO / A The experimental value is 2.15, and the prediction steps based on the QSAR model are as follows:
[0057] logK SiO / A =2.888+0.025×41.038–0.244×6.780=2.26
[0058] The error is only 0.11, which is in good agreement with the experimental value.
Embodiment 2
[0060] Dimethyl disulfide: first check the molecular structure information of dimethyl disulfide on PubChem, and then use the B3LYP / 6-311G** method in the quantum chemistry software Gaussian to calculate α, E LUMO -E HOMO These two descriptors; use the Williams graph method to calculate its h iThe value is 0.031-3, indicating that this compound is within the application domain of the QSAR model constructed in the specific embodiment of the present invention.
[0061] Substituting into the constructed QSAR model, the logK of dimethyl disulfide SiO / A The experimental value is 2.86, and the prediction steps based on the QSAR model are as follows:
[0062] logK SiO / A =2.888+0.025×61.192–0.244×5.736=3.02
[0063] The error is only 0.16, which is in good agreement with the experimental value.
Embodiment 3
[0065] 2-Chlorophenol: first check the molecular structure information of 2-chlorophenol on PubChem, and then use the B3LYP / 6-311G** method in the quantum chemistry software Gaussian to calculate α, E LUMO -E HOMO These two descriptors; use the Williams graph method to calculate its h i The value is 0.109<h*(warning value)=0.25, standard residual (SE)=1.633<3, indicating that this compound is within the application domain of the QSAR model constructed in the specific embodiment of the present invention.
[0066] Substituting into the constructed QSAR model, the logK of 2-chlorophenol SiO / A The experimental value is 4.25, and the prediction steps based on the QSAR model are as follows:
[0067] logK SiO / A =2.888+0.025×85.194–0.244×5.254=3.74
[0068] The error is only 0.51, which is in good agreement with the experimental value.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com