Pseudocercospora rDNA and application thereof to molecular detection of pseudocercospora
A technology for molecular detection and Cercospora, which is applied in the direction of recombinant DNA technology, DNA/RNA fragments, and microbial-based methods, can solve problems such as cross-host infection, and achieve the effects of increasing scope, convenient use, and wide application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] The rDNA full-length sequence of embodiment 1 Pseudomonas mulberry is obtained
[0044] 1. Using the method of high-throughput sequencing, the full-length rDNA of the pathogenic bacteria of mulberry leaf spot disease—Pseudomonas mulberry was obtained by cloning, as shown in SEQ ID NO.1.
[0045] 2. According to the multiple confirmations of the sequencing results, the full-length rDNA of Pseudomonas mulberry was annotated. As shown in the table, the relationship between each gene segment and the transcriptional spacer (ITS) of the rDNA of Pseudomococcus mulberry was determined. Nucleotide sequence and its position on rDNA.
[0046] Table 1 Annotation of the complete rDNA sequence of Pseudomonas sp.
[0047] Features Start(bp) End(bp) Length(bp) ETS1 1 332 332 18S rRNA 333 2059 1726 ITS1 2060 2225 165 5.8S rRNA 2226 2366 140 ITS2 2367 2516 149 28S rRNA 2517 5815 3298 ETS2 5816 6084 268
Embodiment 2
[0048] Embodiment 2 detection primer design and establishment of PCR amplification method
[0049] 1. Primer design
[0050] On the basis of obtaining the full-length rDNA of Pseudomonas mulberry, several pairs of primers were designed. After a large number of specificity and sensitivity tests, two pairs of representative primer sets were finally selected. The primer sequences are as follows:
[0051] P1-F / P1-R Primer Set
[0052] Upstream primer P1-F: 5'-GTTTCAACGGGTAACGGGGA-3'
[0053] Downstream primer P1-R: 5'-TCCCTACCTGATCCGAGGTC-3'
[0054] W1724f / W2196r Primer Set
[0055] Upstream primer W1724f (SEQ ID NO.4): 5'GCTACACTGACAGAGCCAACG 3'
[0056] Downstream primer W2196r (SEQ ID NO.5): 5'GCTACACTGACAGAGCCAACG 3'.
[0057] 2. Establishment of PCR amplification method
[0058] The total DNA of diseased leaves, branches, and soil was used as a template, and the primers described in Example 1 were used for PCR amplification.
[0059] The PCR reaction system (total vol...
Embodiment 3
[0070] Specific detection of embodiment 3 primer W1724f / W2196r
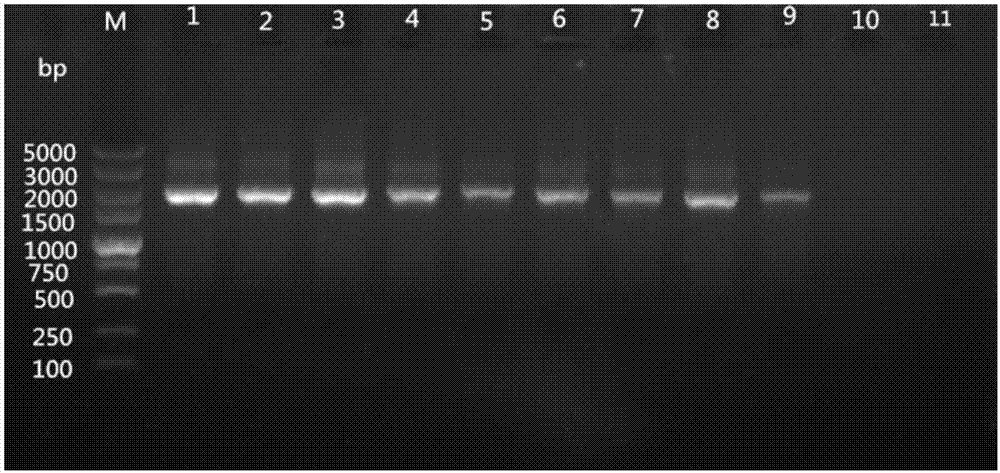
[0071] 1. Separate the fungus and bacteria from the mycelia of stained leaf spot, and the powdery mildew pathogen (Phyllactinia moricola) and the mulberry sclerotinia pathogen (Ciboriacarunculoides) that are the same as the pathogenic fungus of mulberry as contrast group, use primers W1724f / W2196r, carry out PCR amplification with the method of Example 2, agarose gel electrophoresis detection result after amplification.
[0072] 2. The amplification results of the primers are as attached Figure 4 shown. The results showed that only the DNA of the pathogen of mulberry leaf spot disease (Pseudocercia mulberry) had a band at the target position (473bp). It shows that the primer set can specifically detect Pseudomonas mulberry.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com