SNP molecular marker combination for maize genotyping and application thereof
A molecular marker and genotyping technology, applied in the fields of bioinformatics and molecular plant breeding, molecular biology, and genomics, can solve the problems of small quantity, inability to meet large-scale commercial breeding, cumbersome operation process, etc., and achieve reduction Labor cost, accurate and reliable genotyping data, good genetic stability and repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1 is used for the development of the SNP molecular marker combination of maize genotyping
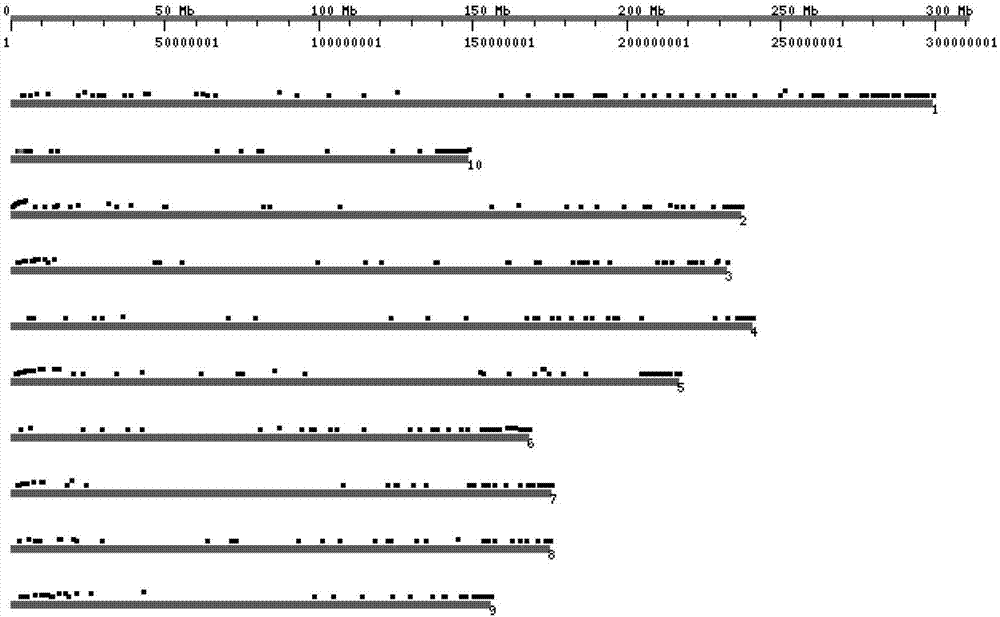
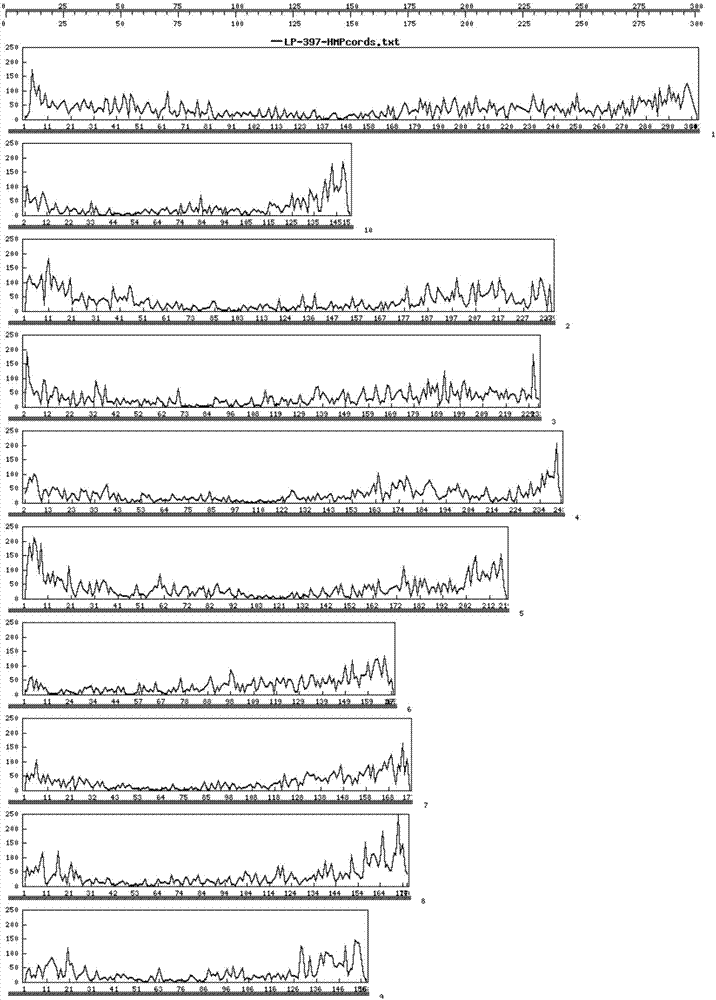
[0031] Whole genome sequencing of 276 maize cultivars was performed using exome sequencing technology. At the same time, the genome sequencing data of 206 maize varieties were sequenced using restriction endonuclease-associated site sequencing technology, and a total of 397 maize materials were sequenced. Combining the resequencing data of other maize varieties in the public database (including the SNP information of 540 maize materials of Professor Yan Jianbing of Huazhong Agricultural University and the SNP information of 503 materials of Hirsch et al. (2014)), the maize B73 genome AGPv3 was used as the reference sequence , analyzed and identified SNP loci, and selected representative, 397 maize materials with a total of 64656 unique SNP markers. Follow the steps below to identify and screen core SNPs.
[0032] 1. Alignment of sequencing reads and maize B73 referen...
Embodiment 2
[0067] Embodiment 2 utilizes KASP technology to detect the method for maize genotype
[0068] According to the core SNP sequence obtained in Example 1, the maize genotype was detected in combination with KASP technology. The specific method is as follows:
[0069] 1. Design KASP primers according to the 50 bp nucleotide sequences flanking each of the 448 SNP sites;
[0070] 2. Select representative corn materials, extract genomic DNA, perform PCR reaction, and verify the availability of KASP primers according to the results of PCR reaction;
[0071] 3. Use the verified KASP primers and SNP high-throughput typing equipment to perform genotyping of the maize material to be tested, so as to identify the genotype of the maize to be tested.
[0072] The sequences of some primers used are listed in Table 2.
[0073] Table 2 partial primer sequence (5'-3')
[0074]
[0075] Step 3 is as follows:
[0076] (1) extracting the genomic DNA of the corn sample to be tested;
[0077...
Embodiment 344
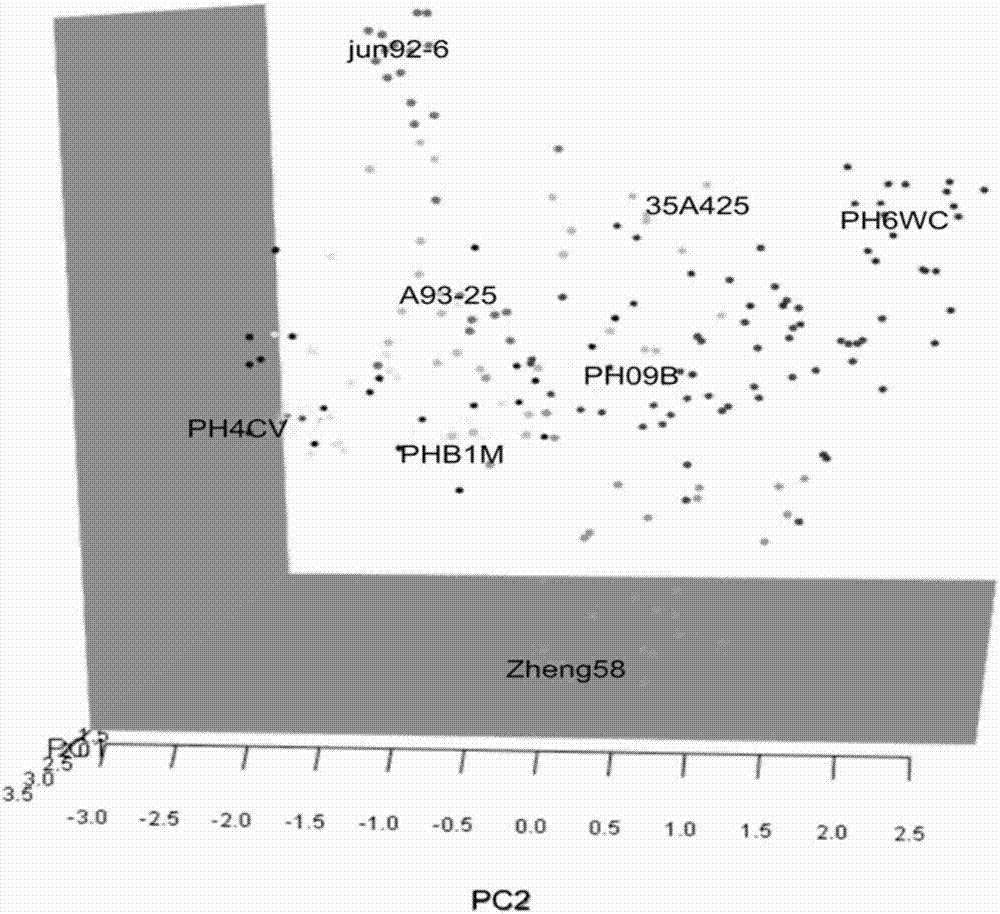
[0083] Example 3 PCA analysis of 448 SNP markers
[0084] For the PCA analysis results of 448 SNP markers, see Image 6 , the paternal group and the maternal group can be clearly divided, which is consistent with the clustering and group structure analysis results.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com