Biosensing detection method for salmon trout seed identification
A technology of biological sensing and detection methods, applied in the field of aquatic organisms, can solve the problems of long detection period, high requirements for detection equipment and operators, and achieve the effect of short response time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Embodiment 1 identifies whether rainbow trout fry is infected with IHNV
[0064] 1. Solution preparation
[0065] Custom-synthesized primers with base sequences as shown in SEQ ID NO: 1-6 (Shanghai Shenggong), each primer has a ferrocene modification at the 5' end, dissolved in nuclease-free sterilized deionized water, and prepared to contain 1 μM each of primers SEQ ID NO: 1 and SEQ ID NO: 2, 8 μM each of primers SEQ ID NO: 3 and SEQ ID NO: 4, and 2 μM each of primers SEQ ID NO: 5 and SEQ ID NO: 6. Primers SEQ ID NO: 3 and SEQ ID NO: 4 have a ferrocene tag upstream.
[0066] Use nuclease-free sterilized deionized water to prepare a dNTP use solution, which contains 10 mM each of dATP, dCTP, and dGTP, 8 mM of dTTP, and 2 mM of biotin-16-dUTP.
[0067] 2. Total RNA Extraction
[0068] The body length of the rainbow trout fry to be tested is about 2-3cm. Dissect 5 tails, take the viscera and mix them, grind them with liquid nitrogen, use the RNAsimple Total RNA Extract...
Embodiment 2
[0084] Embodiment 2 Identify whether golden trout eggs carry IHNV
[0085] 1. Solution preparation
[0086] Custom-synthesized primers with base sequences as shown in SEQ ID NO: 1-6 (Shanghai Sangong), the 5' ends of primers SEQ ID NO: 1-4 are all modified with ferrocene, dissolved in nuclease-free sterilized Deionized water, formulated to contain 1 μM each of primers SEQ ID NO: 1 and SEQ ID NO: 2, 8 μM each of primers SEQ ID NO: 3 and SEQ ID NO: 4, 2 μM each of primers SEQ ID NO: 5 and SEQ ID NO: 6 The primer using solution, wherein primers SEQ ID NO: 3 and SEQ ID NO: 4 are labeled with ferrocene upstream.
[0087] Use nuclease-free sterilized deionized water to prepare a dNTP use solution, which contains 10 mM each of dATP, dCTP, and dGTP, 8 mM of dTTP, and 2 mM of biotin-16-dUTP.
[0088] 2. Total RNA Extraction
[0089] Take about 20 pieces of golden trout eggs to be tested, mix them, grind them with liquid nitrogen, use the RNAsimple Total RNA Extraction Kit (Tiangen D...
Embodiment 3I
[0102] The quantitative detection of embodiment 3IHNV
[0103] 1. Solution preparation
[0104] With embodiment 1.
[0105] 2. IHNV control plasmid extraction
[0106] According to the whole genome sequence (KJ421216.1) of IHNV Chinese strain Ch20101008 in NCBI database, the synthetic matrix (matrix, M) gene and its 10 bp sequences (2245-2852) at both ends of the flanks, a total of 608 bp, were connected to the pGM-T vector, An IHNV control plasmid with a molecular weight of 3639bp was constructed, transformed into DH5αEscherichia coli, and expanded for culture. Use the endotoxin-free plasmid medium extraction kit (Tiangen DP108) to extract the plasmid, dilute it in a 10-fold concentration gradient, and prepare it to contain 1-10 13 A solution of plasmid copies.
[0107] 3. Nucleic Acid Amplification
[0108] The amplification reaction system is as follows:
[0109]
[0110] Each reaction tube was placed in a 60 °C water bath for 30 min.
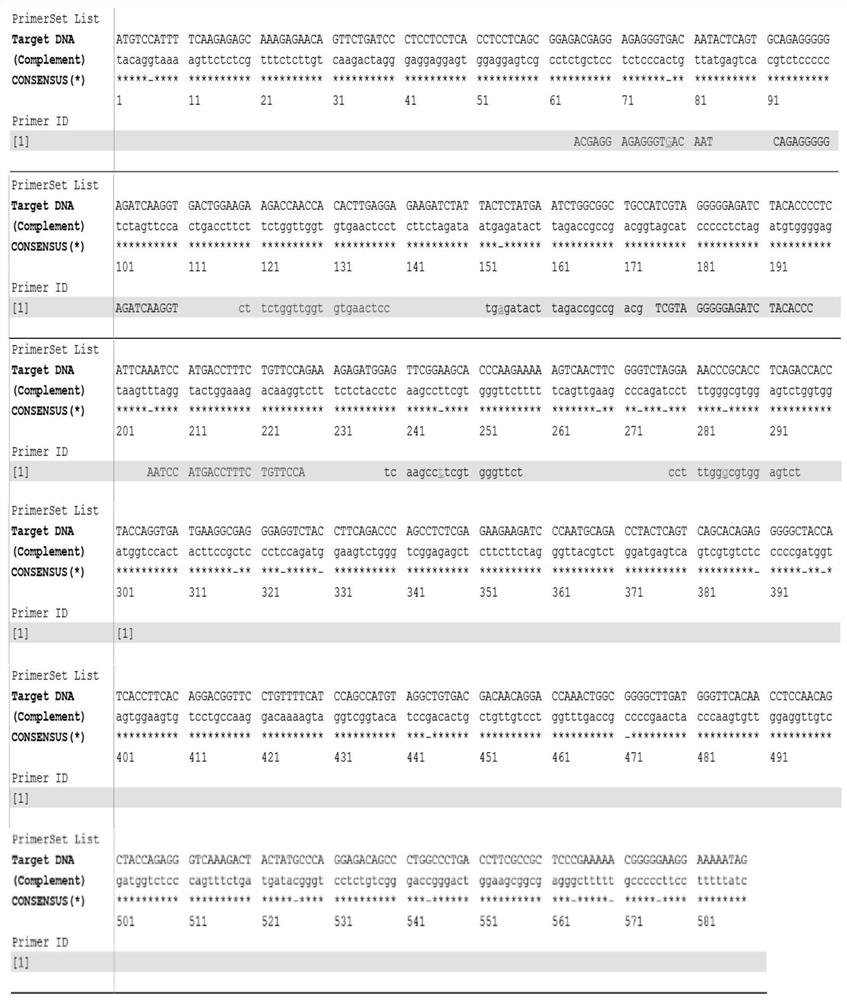
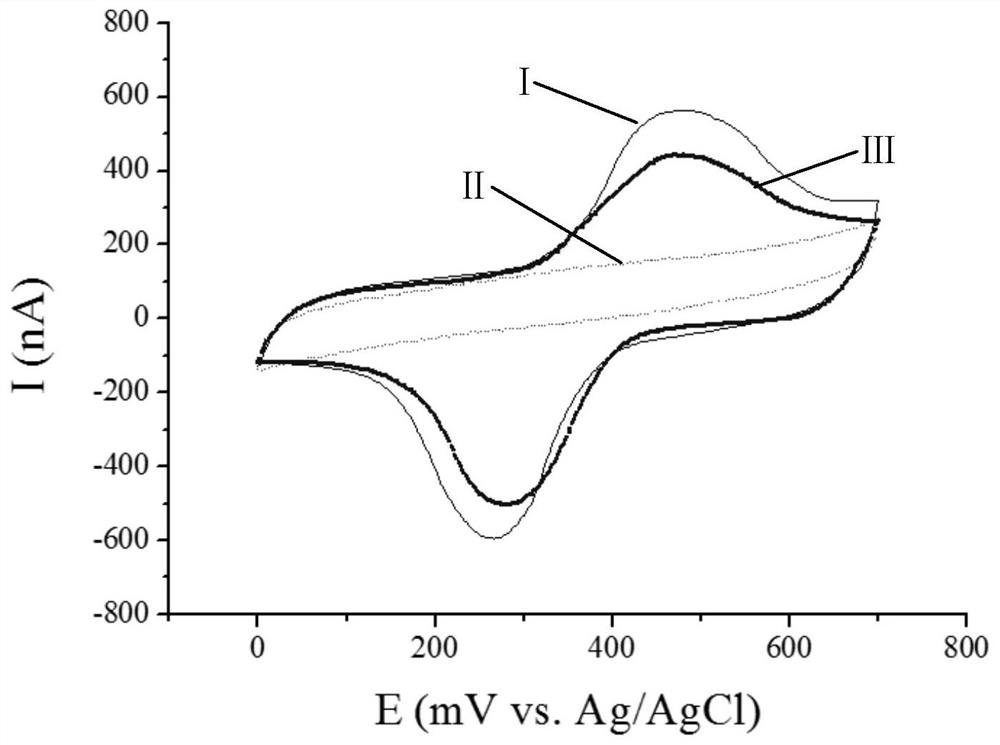
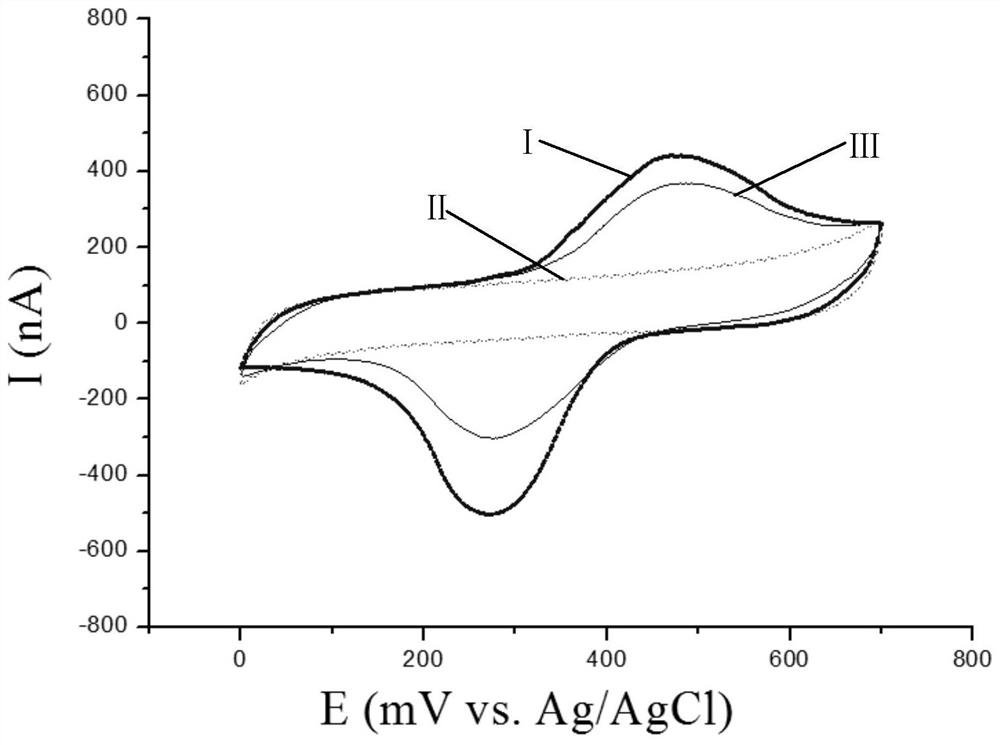
[0111] 4. Electrode Surface...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com