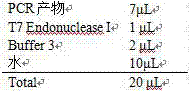Method for increasing non-homologous end joining efficiency of CRIPSR/Cas9 target knock-out genes
A gene and efficiency technology, applied in the field of improving the efficiency of non-homologous end joining of CRIPSR/Cas9 targeted knockout genes, can solve the problems of reducing the fidelity of the repair process and gene breakage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0013] The preferred embodiments of the present invention will be described in detail below with reference to the accompanying drawings. For the experimental methods that do not specify specific conditions in the examples, generally follow the conventional conditions, such as the conditions described in the Molecular Cloning Experiment Guide (Third Edition, J. Sambrook et al.), or the conditions suggested by the manufacturer.
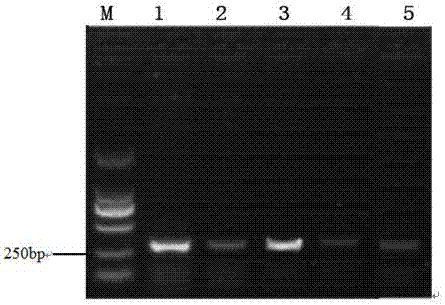
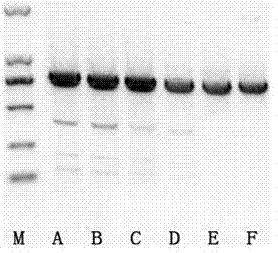
[0014] According to the NCBI database, the CDS region of the gene PTEN was discovered, and the primer sequence was designed in combination with NCBI Primer-BLAST. At the same time, according to the patent (201611252179.5), the primer for the amplification of the target site region of the lin28a gene was synthesized. The sequence is as follows:
[0015]
[0016] Five small interfering RNA sequences targeting PTEN gene were designed and synthesized by online software, and the corresponding RNA sequences are shown in SEQ ID NO.1--SEQ ID NO.5.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com