Whole genome copy number detection chip customized for X chromosome high-density probe
A detection chip and whole genome technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of low resolution of CNV screening, inability to be detected, low probe density, etc., and improve genetic diagnosis Ability, high degree of automation, easy to promote the effect of application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1: Design principle and preparation steps of the chip of the present invention
[0027] 1. Chip probe design:
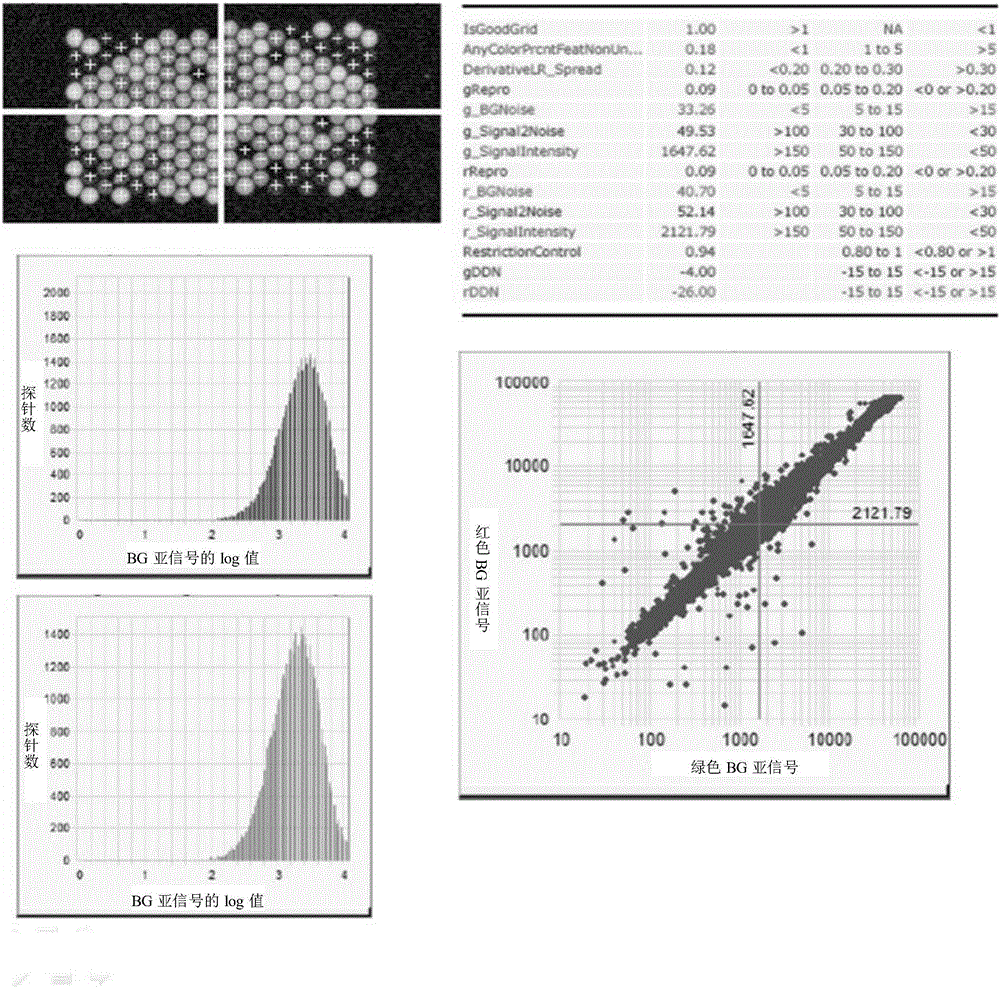
[0028] Firstly, 30,000 probes in the probe library (https: / / earray.chem.agilent.com / suredesign / ) were tiled in the whole gene. At this time, there were only 2509 probes on the X chromosome. After that, 18,837 probes were added for 90 important disease-related genes on the X chromosome. The specific design was as follows: first, 10,000 probes were tiled for the 90 genes on the X chromosome, and then 1,502 exomes of 90 genes were selected. 9712 probes were designed for the sub-region and its adjacent region 200bp (100bp each on the left and right), among which, more than 98% of the exons were designed with at least 2 probes, and more than 88% of the exons were designed with at least 3 probes, More than 69% of the exons were designed with at least 4 probes, and more than 44% of the exons were designed with at least 5 probes (see Table 1 below for spec...
Embodiment 2
[0055] Embodiment 2: detection embodiment of the chip of the present invention
[0056] (1) Whole-genome DNA extraction of the sample to be tested: it can be extracted according to conventional methods in the art, and the sample can be human venous anticoagulated blood. The present invention adopts a commercial DNA extraction kit (Qiagen, QIAamp DNA Blood Mini Kit, CatNo.51104 ) for genome-wide DNA extraction; use Nanodrop 2000, Qubit 2.0 for DNA quality and concentration detection.
[0057] (2) Digestion of the control sample and the sample to be tested (Agilent, SureTag DNA Labeling Kit, CatNo.5190-3399): Take 0.2-0.5ug of genomic DNA from each of the sample to be tested and the control sample, place them in different Ep tubes, and dilute with water To 10.1ul, the sex of the control sample is the same as that of the sample to be tested (control sample Agilent, Cat No.5190-4370, 5190-4371), according to the 2.9ul system, add the following mixed enzyme digestion solution (incl...
Embodiment 3
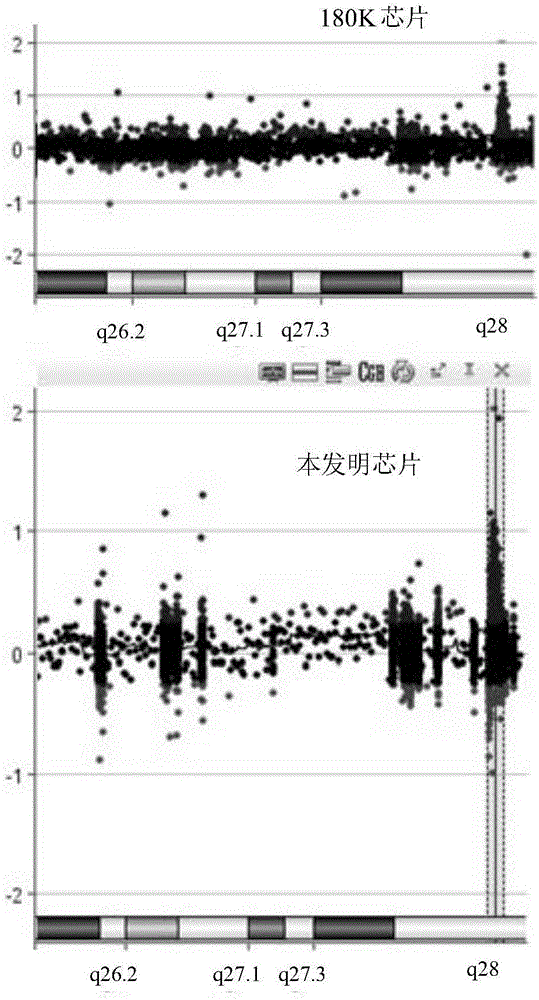
[0067] Embodiment 3: Two comparative detection embodiments of the chip of the present invention and the existing chip
[0068] 1. MECP2 Microduplication Syndrome
[0069] The existing SurePrint G3 180K chip (Agilent, Cat. G4884A) and the chip of the embodiment of the present invention were used to detect DNA samples of patients with MECP2 microduplication syndrome according to the method of Example 2. from figure 2It can be seen that in the detection results of the chip of the present invention, an abnormal peak (at the dotted line) can be clearly seen in the Xq28 area, and the position of the abnormal peak is basically the same as that of the 180K chip, but its peak is obviously higher than that of the 180K chip. Experimental results have confirmed that the 60K chip of the present invention can accurately detect MECP2 microduplication syndrome. Since the price of the 60K chip is lower than that of the commercial 180K chip, if the customized 60 chip is used, the economic pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com