Primer combination for simultaneously identifying 8 kinds of cattle pathogens and GeXP detection method
A combination of primers and pathogens technology, applied in the direction of microbial-based methods, biochemical equipment and methods, microbial measurement/testing, etc., can solve problems such as incorrect results, time-consuming and labor-intensive problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
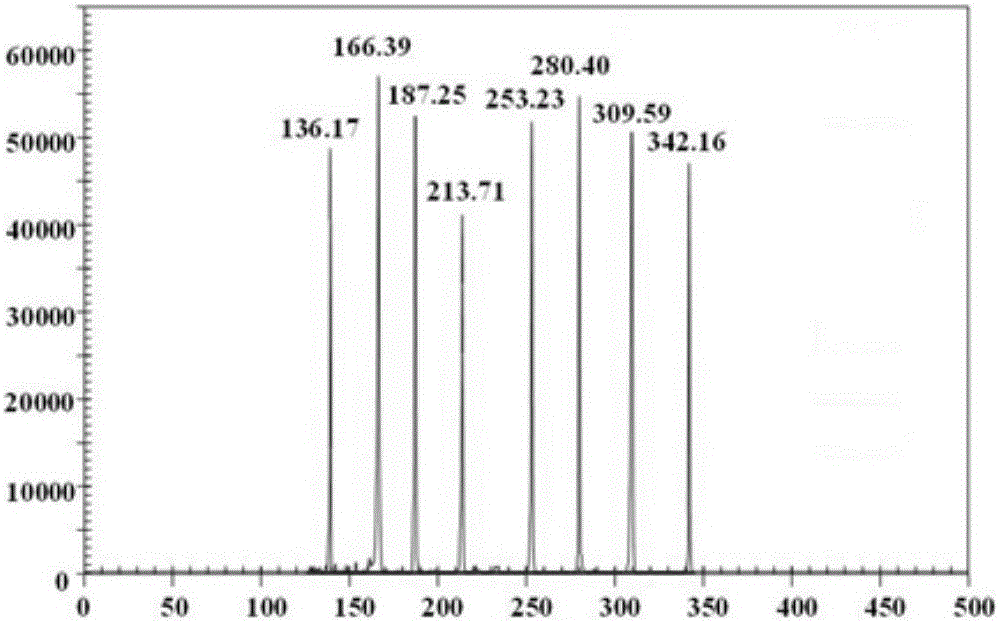
[0153] Embodiment 1, design and preparation of primer combination
[0154] A large number of sequence analyzes and comparisons were carried out to obtain several primers for identifying 8 bovine pathogens of FMDV, BTV, VSV, BVDV, BRV, ETEC, IBRV and PPRV. Preliminary experiments were carried out on each primer to compare performances such as sensitivity and specificity, and finally 8 pairs of specific primers for identifying 8 kinds of bovine pathogens were obtained. Each specific primer pair, forward primer and reverse primer, consists of a targeting segment and a universal primer segment, with the universal primer segment located 5' to the targeting segment.
[0155] The primer pair used to identify FMDV consists of the following two primers (5'→3'):
[0156] FMDV-F (Sequence 1 of the Sequence Listing): AGGTGACACTATAGAATA GCCGTGGGACCATACAGG;
[0157] FMDV-R (Sequence 2 of the Sequence Listing): GTACGACTCACTATAGGGA AAGTGATCTGTAGCTTGGAATCTC.
[0158] The underlined part...
Embodiment 2
[0196] Embodiment 2, specificity
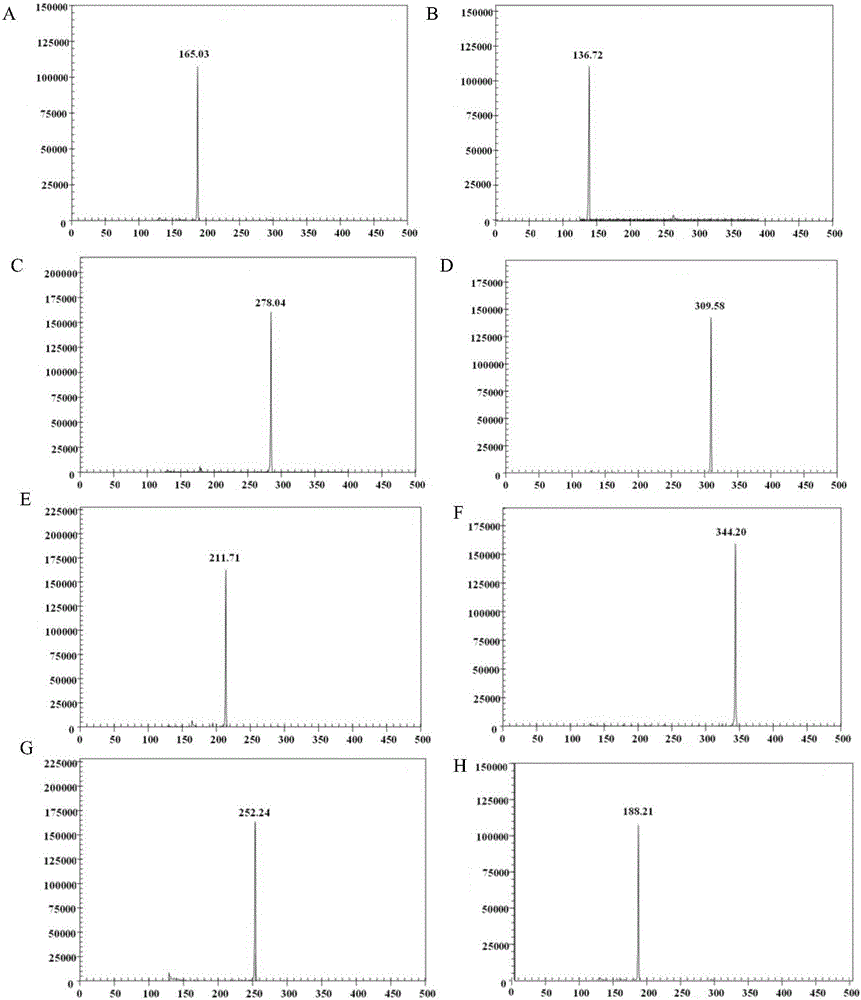
[0197] 1. Single template experiment
[0198] 1. Extract the total RNA of the sample to be tested and reverse transcribe it into cDNA. The samples to be tested are: FMDV type O inactivated virus, BTV type 4 inactivated virus, VSV NJ type inactivated virus, BVDV reference strain Oregon CV24 strain (BVDV-1 type), BRV reference strain NCDV, PPRV vaccine strain , IBRV virus.
[0199] 2. Extract the genomic DNA of the sample to be tested. The samples to be tested were ETEC reference strain 1676.
[0200] 3. Using the cDNA obtained in step 1 and the genomic DNA obtained in step 2 as templates, GeXP multiplex PCR was performed using the primer combination in Example 1.
[0201] Multiplex PCR reaction system (20 μL): template 1 μL, Genome Lab GeXP Starter Kit 5×buffer 4 μL (the buffer contains universal primers, and the universal primers are primer A shown in sequence 25 of the sequence listing and primers shown in sequence 26 of the sequence lis...
Embodiment 3
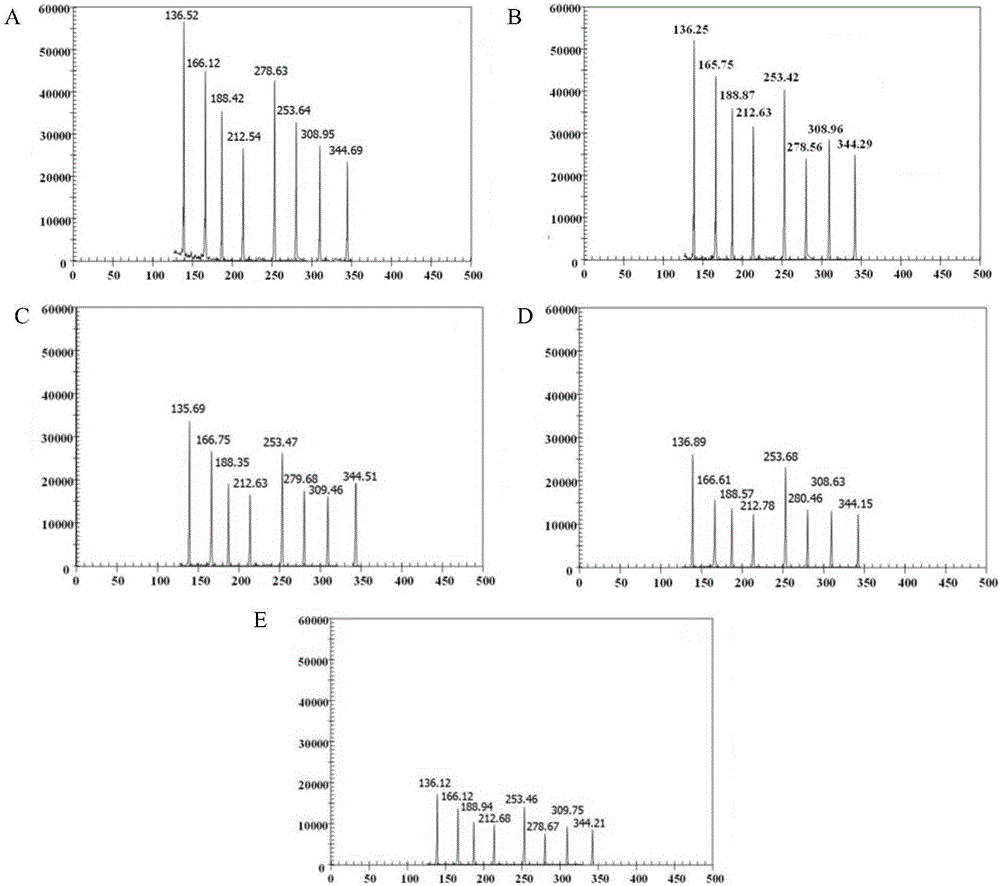
[0219] Example 3, universality
[0220] 1. Extract the total RNA of the sample to be tested and reverse transcribe it into cDNA. The samples to be tested are: FMDV type O inactivated virus, FMDV type A inactivated virus, FMDV Asia type I inactivated virus, BTV type 4 inactivated virus, BTV type 8 inactivated virus, BTV type 9 inactivated virus, BTV Type 15 inactivated virus, BTV type 17 inactivated virus, BTV type 18 inactivated virus, VSV NJ type inactivated virus, VSV IND type inactivated virus, BVDV reference strain Oregon CV24 strain (BVDV-1 type), BVDV reference strain Strain NADL strain (BVDV-1 type), BVDV reference strain Yak strain (BVDV-1 type), BVDV strain GX-BVDV1, BVDV strain GX-BVDV2, BVDV strain GX-BVDV3, BVDV strain GX- BVDV4, BVDV strain GX-BVDV5, BVDV strain GX-BVDV6, BVDV strain GX-BVDV7, BVDV strain GX-BVDV8, BVDV strain GX-BVDV9, BVDV strain GX-BVDV10, BVDV strain GX-BVDV11 , BVDV strain GX-BVDV12, BVDV strain GX-BVDV13, BVDV strain GX-041, BRV reference ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com