Method and device for determining microbial species
A microbial species, microorganism technology, applied in the field of microbial detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
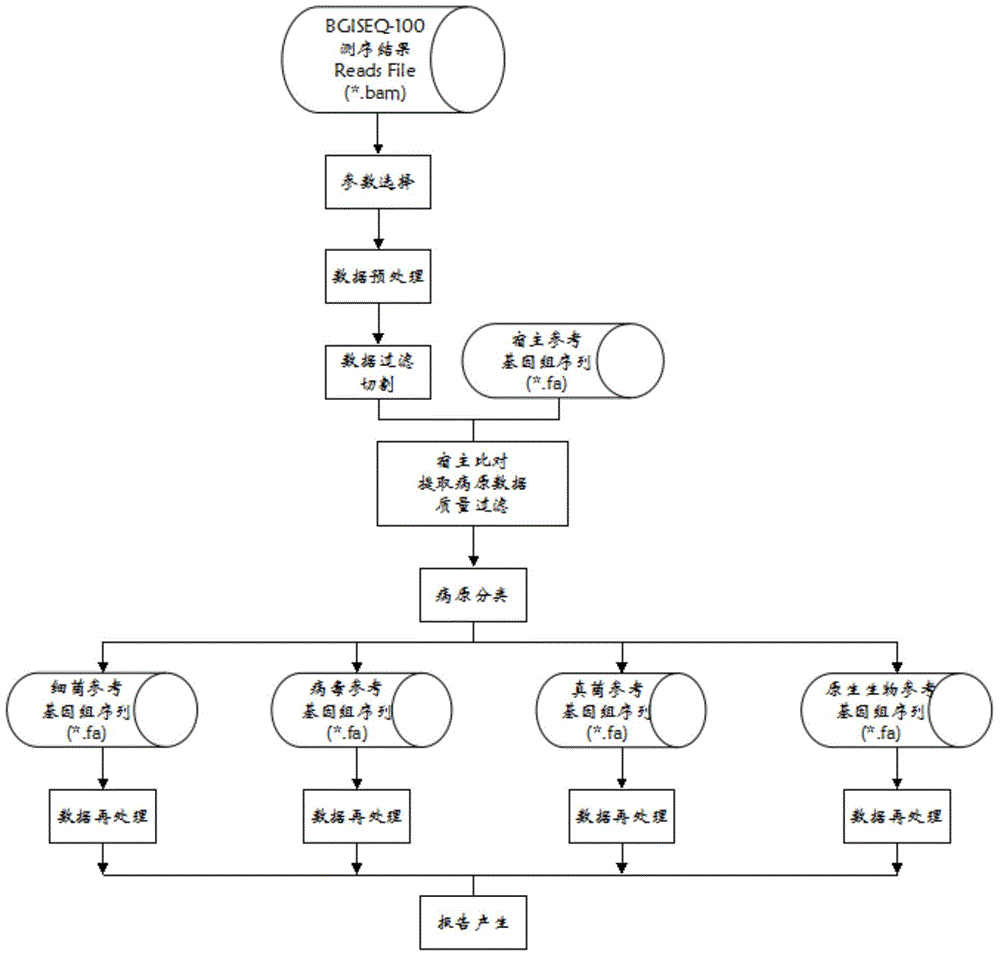
[0053] A urine sample (sample name: UPUR14GY0001) of a clinical patient was obtained from the hospital, data preprocessing was performed on the data generated by sequencing, and the generated data was converted into the standard FASTQ format. It is then aligned to the host's reference genome, and those sequences that do not align and those whose similarity threshold is below the specified threshold are extracted. Then compare the extracted sequences to the four database files of bacteria, fungi, viruses, and protists to classify pathogenic microorganisms. Then each comparison result (comparison file) is deduplicated, coverage and other statistical values are calculated, and finally a readable result file is generated.
[0054] The steps and processes of this detection system are integrated into a software package named UPMD (Unknown Pathogen Microorganism Detection). The operating environment of this software is the Unix / Linux operating system, and it runs through the Unix / L...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com