Detecting method for food-borne pathogenic bacteria listeria monocytogenes
A technology for the detection of Listeria monocytogenes and its purpose, which is applied in the field of detection of food-borne pathogenic bacteria Listeria monocytogenes, can solve the problems of long inspection cycle, inability to be widely used and popularity in the market, and achieve simple detection , effectively the occurrence of listeriosis monocytogenes epidemic, and the effect of inhibiting the occurrence of listeriosis monocytogenes epidemic
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Embodiment 1: Design and optimization of primers and probes
[0025] Step 1) carry out primer design according to the hly gene (hlyA) of the encoding hemolysin O (listeriolysinO) of Listeria monocytogenes:
[0026] The public listeriolysinO coding gene hly was found through NCBI, and the site of its conserved region was the 1657-3060 position of the hly gene. After homology analysis, RPA primers were designed with the gene located at 1645-3069 as the target fragment.
[0027] Design 4 sets of primer pairs for optimal primer screening, the primer sets and sequences are as follows:
[0028] Group 1:
[0029] Forward primer F1-RPA: 5′-CCGTAAGTGGGAAATCTGTCTCAGGTGATGTAG-3′(33bp)
[0030] Reverse primer R1-RPA: 5′-TTGTCTTTTTAAGAAGTTTGTTGTATAGGCAATGGG-3′(35bp)
[0031] Product length: 220bp
[0032] Group 2:
[0033] Forward primer F2-RPA: 5′-CTTTTGACGCTGCCGTAAGTGGGAAATCTGT-3′(31bp)
[0034] Reverse primer R1-RPA: 5′-TTGTCTTTTTAAGAAGTTTGTTGTATAGGCAATGGG-3′(35bp)
[0035...
Embodiment 2
[0053] Embodiment 2: the detection of primer and probe effect
[0054] Establishment of Isothermal Amplification Method for Recombinase Polymerase
[0055] Include the following steps
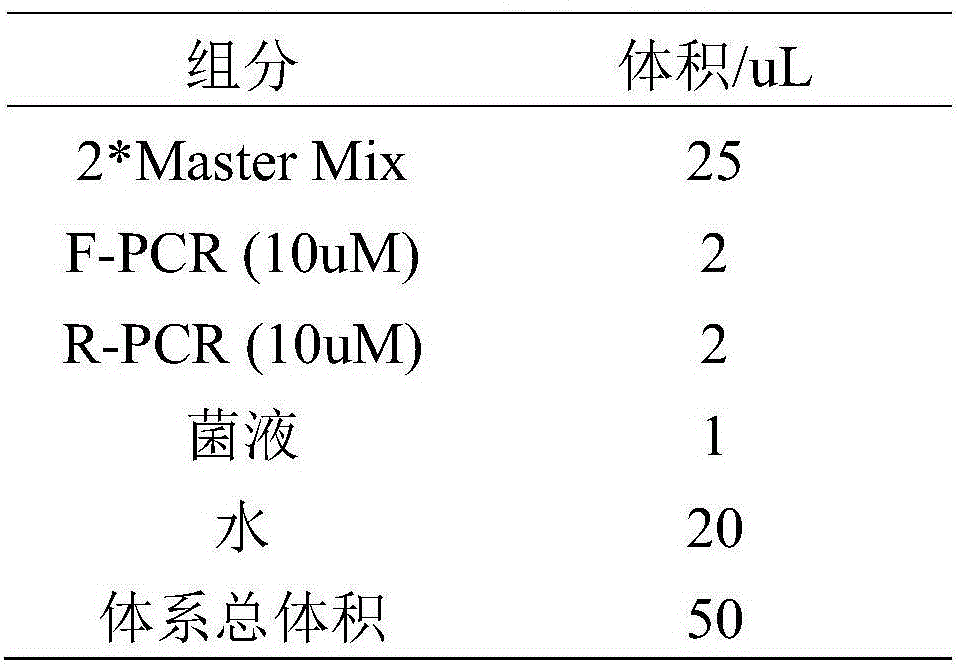
[0056] 1) Prepare the RPA reaction system:
[0057] The final concentration of each primer is: primer F-RPA and primer R-RPA each 420nmol / μL; probe P120nmol / μL; buffer composition and concentration are: Tris-HCl (pH7.9) 50mmol / μL, KAc (potassium acetate ) 100mmol / μL, DTT (dithiothreitol) 2mmol / μL, 5% PEG20mol / μL, dNTPs200umol / μL, ATP3mmol / μL, pcr (phosphocreatine kinase) 50mmol / μL, CK (creatine kinase) 100ng / μL, DNA polymerase Bsu30ng / uL, single-strand binding protein Gp32300ng / uL, recombinase UvsX240ng / uL, auxiliary enzyme UvsY60ng / uL, endonuclease Nfo200ng / uL; sample DNA template 10ng, add double distilled water to make unreacted The volume of the reaction mix is 47.5 μL; 280 mmol / μL MgAc (magnesium acetate) is 2.5 μL, so that the total volume of the reaction system is 50 μL. Magnesium...
Embodiment 3
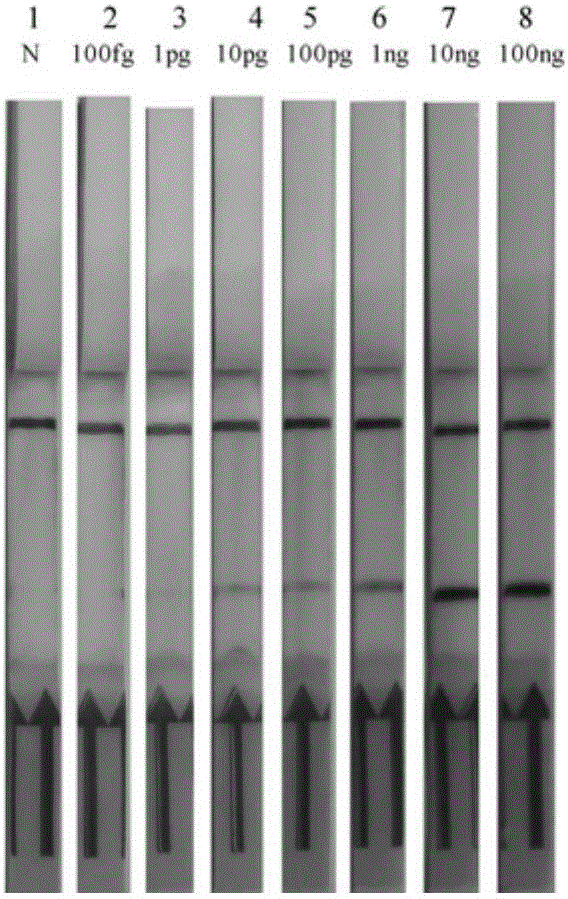
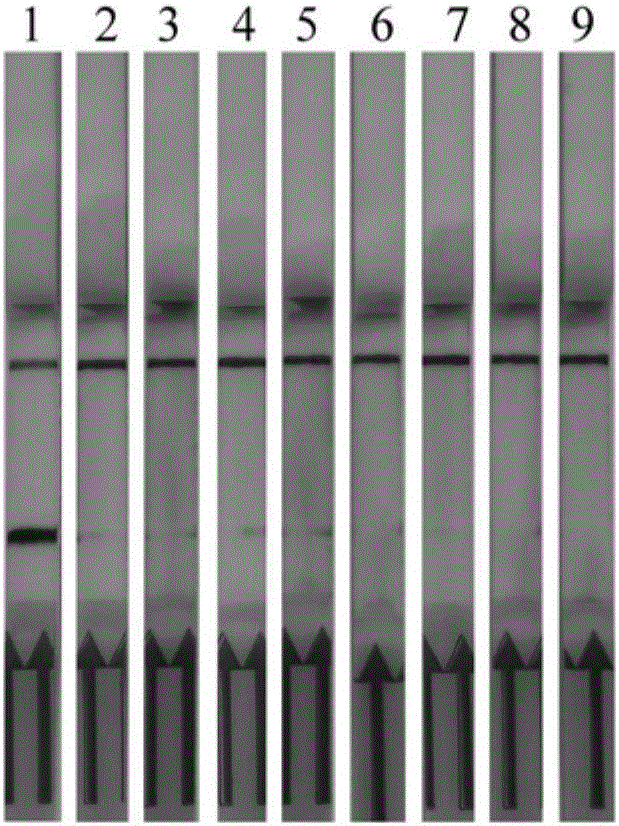
[0068] Example 3: Application of RPA-LFD to the detection of artificially polluted samples
[0069] 3.1 Materials: pork (P), chicken (C), fish (B), beef (F); Mengniu pure milk (M). All materials were purchased from supermarkets.
[0070] 3.2 Reagents: Brain Heart Infusion (BHI) sterile liquid medium, sterile water (nuclease-free)
[0071] 3.3 Sample handling:
[0072] 3.3.1 Meat
[0073] After peeling and bone removal, wash with sterile water, cut into micro pieces, weigh 50-100mg, add 400uL pre-cooled sterile double-distilled water, grind manually with a tissue grinder to make a homogenate. Centrifuge at 4000rpm for 15min, discard the upper layer of fat layer and the lower layer of sediment, take the middle layer, pass through the membrane at 0.22um, and sterilize.
[0074] 3.3.2 Milk
[0075] Take an appropriate amount of milk, dilute it 50 times with sterile double distilled water, lyse it directly, pass it through a membrane at 0.22um, and sterilize.
[0076] 3.4 Sam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com