Primer, method and application based on next-generation sequencing technology to detect HBV drug-resistant mutation site
A technology for the detection of drug resistance mutation sites and techniques, which is applied in the fields of genomics and molecular biology, and can solve the problems of difficult separation, insignificant differences in fragment lengths, and large numbers, and achieves the effect of high stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] A method for detecting HBV drug-resistant mutation sites based on next-generation sequencing technology, a round of PCR comprises the following steps:
[0058] 1. Design and synthesize the primers shown in SEQUENCE NO.1 and SEQUENCE NO.2;
[0059] 2. Amplify the HBV RTDNA fragment (HBV552~HBV992), including 10 drug resistance mutation sites
[0060] Configure the PCR system as follows (the DNA template is DNA extracted from 200ul serum of HBV patients, and the extraction kit uses the bloodminikit of qiagen):
[0061]
[0062] The configured system was subjected to PCR reaction according to the following procedures:
[0063]
[0064]
[0065] After the PCR reaction, use 1.5% agarose gel electrophoresis to verify the size of the enriched fragment of the PCR product, select the 441bp target fragment to recover by gel cutting, and purify to 34 μL. The purification steps are as follows:
[0066] 1) Add 3 times the volume of BufferQG (conventional DNA recovery buff...
Embodiment 2
[0076] A method for detecting HBV drug-resistant mutation sites based on next-generation sequencing technology, the steps of a round of PCR include: except that the PCR program is different from that of Example 1, other steps are the same as in Example 1, wherein the steps of this Example 2 The PCR program is as follows:
[0077]
[0078] Example 1 and Example 2 used the same primer pair.
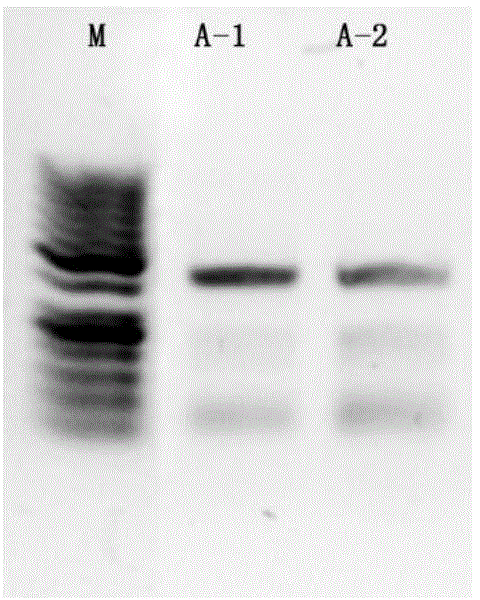
[0079] After the PCR product was purified, an equal volume of the product was taken for electrophoresis, and the results were as follows: figure 1 shown. exist figure 1 Among them, A-1 is the result after purification of the PCR product of Example 1, and A-2 is the result after purification of the PCR product of Example 2. Depend on figure 1 It can be seen that the effect of A-1 is better than that of A-2 in terms of specificity and product quantity.
Embodiment 3
[0081] A method for detecting HBV drug-resistant mutation sites based on next-generation sequencing technology, the steps of one round of PCR include: except that the PCR primer pair is different from that of Example 1, other steps (i.e. conditions such as a round of PCR reaction system and procedures) are all Same as embodiment 1, wherein, the PCR primer pair of present embodiment 3 is as shown in SEQUENCENO.3 and SEQUENCENO.4 as follows:
[0082] SEQUENCE NO.3: CHTGTTGCTGTACAAAACCT
[0083] SEQUENCE NO.4: GCAGGATAWCCACATTG
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com