Method for detecting 5-methylcytosines in nucleic acid
A technology of methylcytosine, nucleic acid, applied in the field of nucleic acid chemistry
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
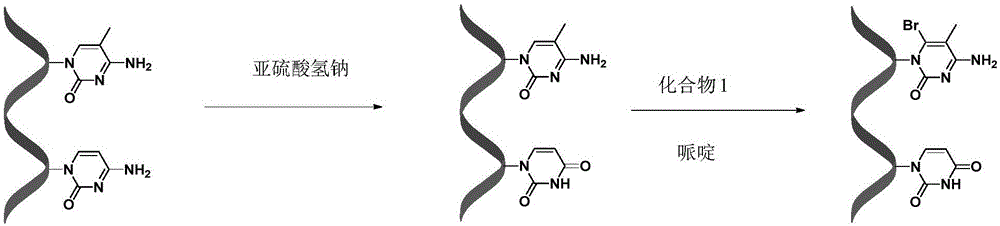
[0023] Example 1: Sodium bisulfite treatment of nucleic acid strands containing 5-methylcytosine
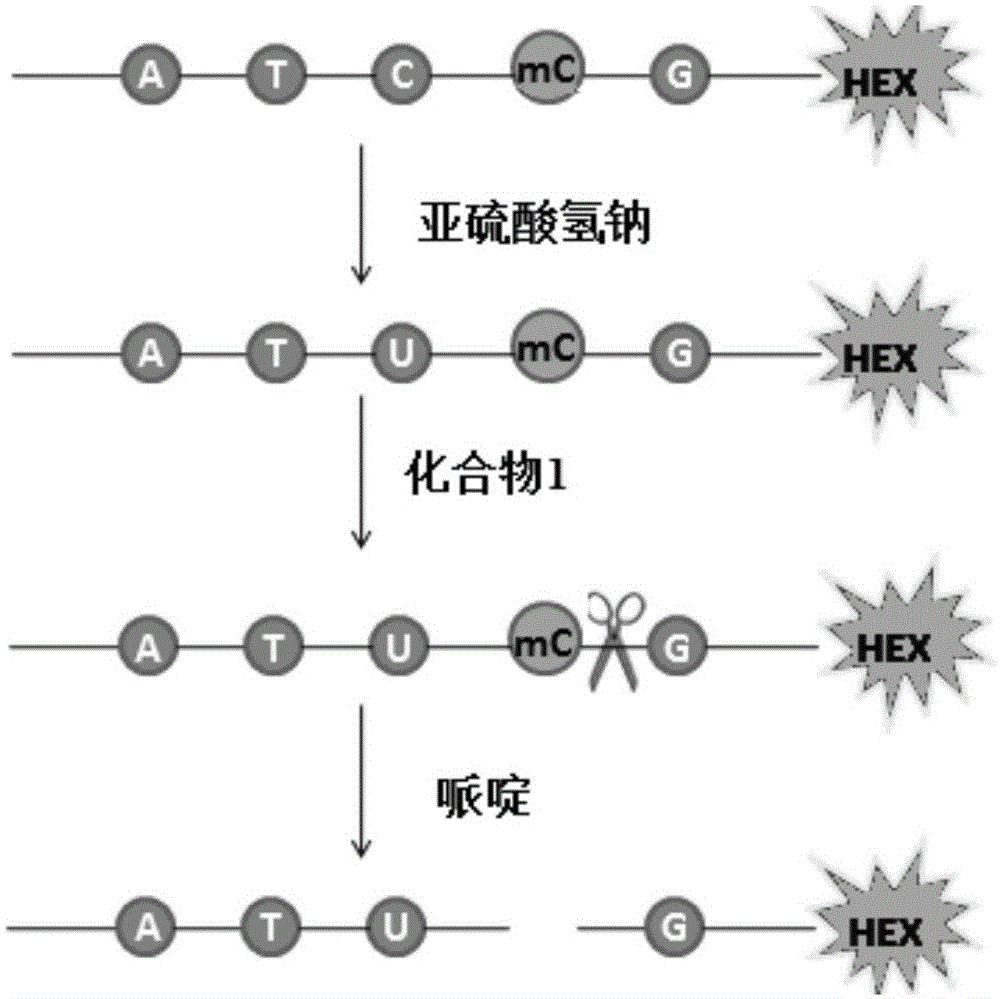
[0024] Pipette 2 μL of 5-methylcytosine-containing DNA (10 μM) labeled with the fluorescent substance HEX (Chinese name is 5-hexachlorofluorescein phosphoramidate) at the 3’ end, add 48 μL of water, and then add 5.5 μL of freshly prepared NaOH solution (3M), incubate in a water bath at 42°C for 0.5 hours, add 30 μL of hydroquinone (10 mM) after the completion of the water bath incubation, and then add 520 μL of NaHSO 3 solution (3.6M, pH=5.0) and 200 μL of paraffin oil, and incubated in a water bath at 50° C. for 16 hours in a dark place, to change cytosine on DNA to uracil. After the reaction, use a desalting column to desalt and purify the DNA.
Embodiment 2
[0025] Example 2: DNA treated with sodium bisulfite was treated with compound 1 and volume fraction of 10% piperidine
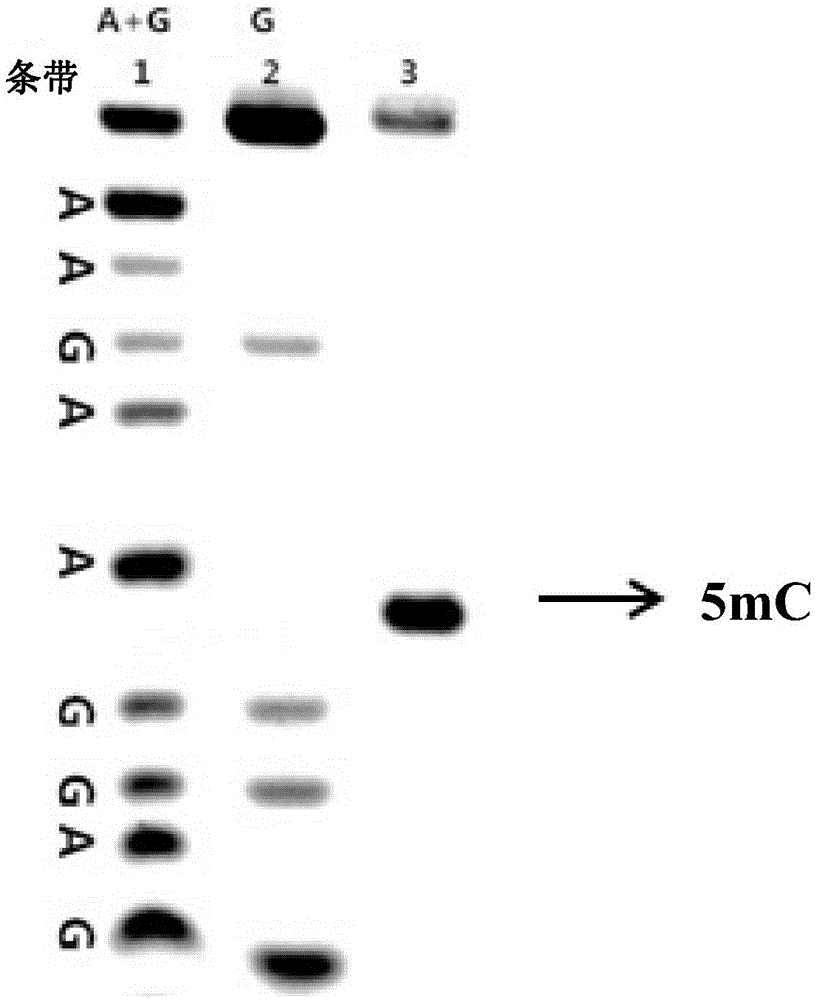
[0026] Take 4 μL of 5-methylcytosine-containing DNA (10 μM) treated with sodium bisulfite, add 10 μL of acetonitrile, 2 μL of Tris-HCl solution (1M, pH=5.0), 2 μL of water and 2 μL of compound 1 ( 20mM), heat treatment at 50°C for 10 minutes, 5-methylcytosine on the DNA chain specifically binds to compound 1; then precipitated with ice ethanol, centrifuged, spin-dried the solvent, and then added piperidine with a volume fraction of 10% at 90°C for 0.5 h, the position-specific cleavage of the 5-methylcytosine bound to compound 1; then ice-ethanol precipitation, centrifugation, and spin-drying of the solvent. Load the sample to the electrophoresis tank, and observe the gel results after 2 hours. By comparing with the control of the A+G sequence and the control of the G sequence, the position and number of the DNA bands in the polyacrylamide denaturing gel can b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com