Mutant of penicillin G acylase (PGA) and preparation method and application of mutant
A technology of penicillin and acylase, which is applied in the field of penicillin G acylase mutants and its preparation, which can solve the problems of low catalytic activity, inability to increase 6-APA production rapidly, long reaction time, etc., and achieve good conversion rate, The effect of fast reaction rate and low substrate residue
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
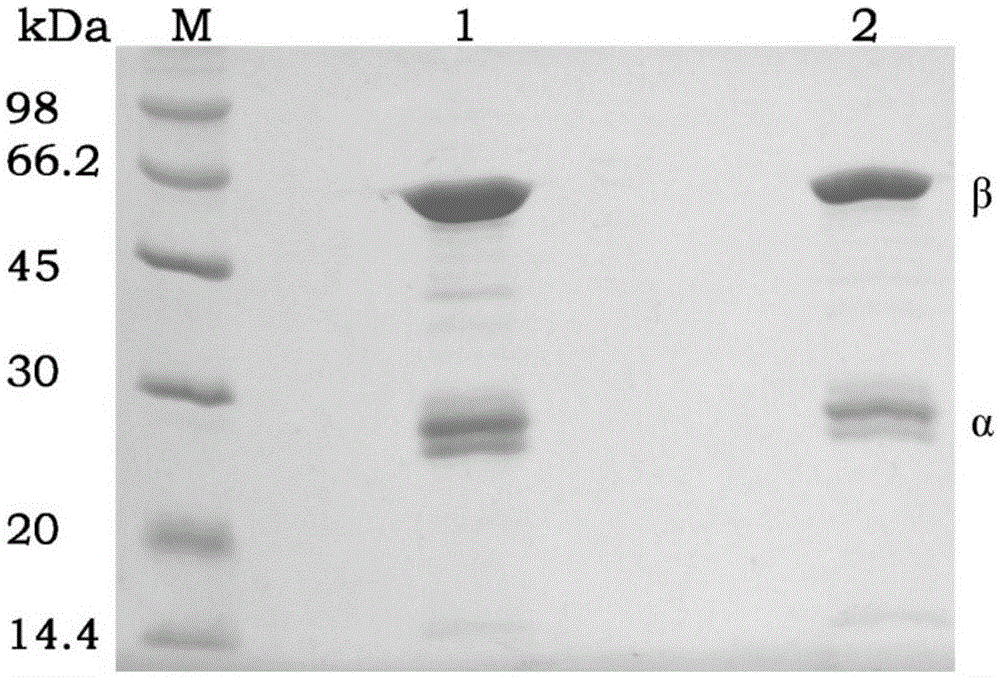
[0063] Example 1: Cloning and expression of wild-type PGA gene and purification and immobilization of recombinant protein
[0064] 1-1 Obtaining of wild-type PGA gene
[0065] According to the amino acid and nucleotide sequence of Escherichia coli ATCC11105 penicillin G acylase provided by GeneBank (GeneBank accession number: X04114.1), the inventors obtained a complete intracellular The mature peptide of PGA (SEQ ID NO: 2) was expressed, and the optimized amino acid was subjected to whole gene synthesis (SEQ ID NO: 1).
[0066] 1-2 Construction of wild-type PGA prokaryotic expression vector
[0067] Primers were designed according to the prokaryotic expression vector pET30a(+) and the wild-type PGA gene synthesized by the whole gene. The specific primer sequences are as follows:
[0068] P1: 5'-CCC AAGCTT ATGGAGCAGTCGTCAAGT-3' (where the base in the underline is the HindIII restriction site)
[0069] P2: 5'-CCG CTCGAG TTATCTCTGAACGTGCAA-3' (the underlined base is the X...
Embodiment 2
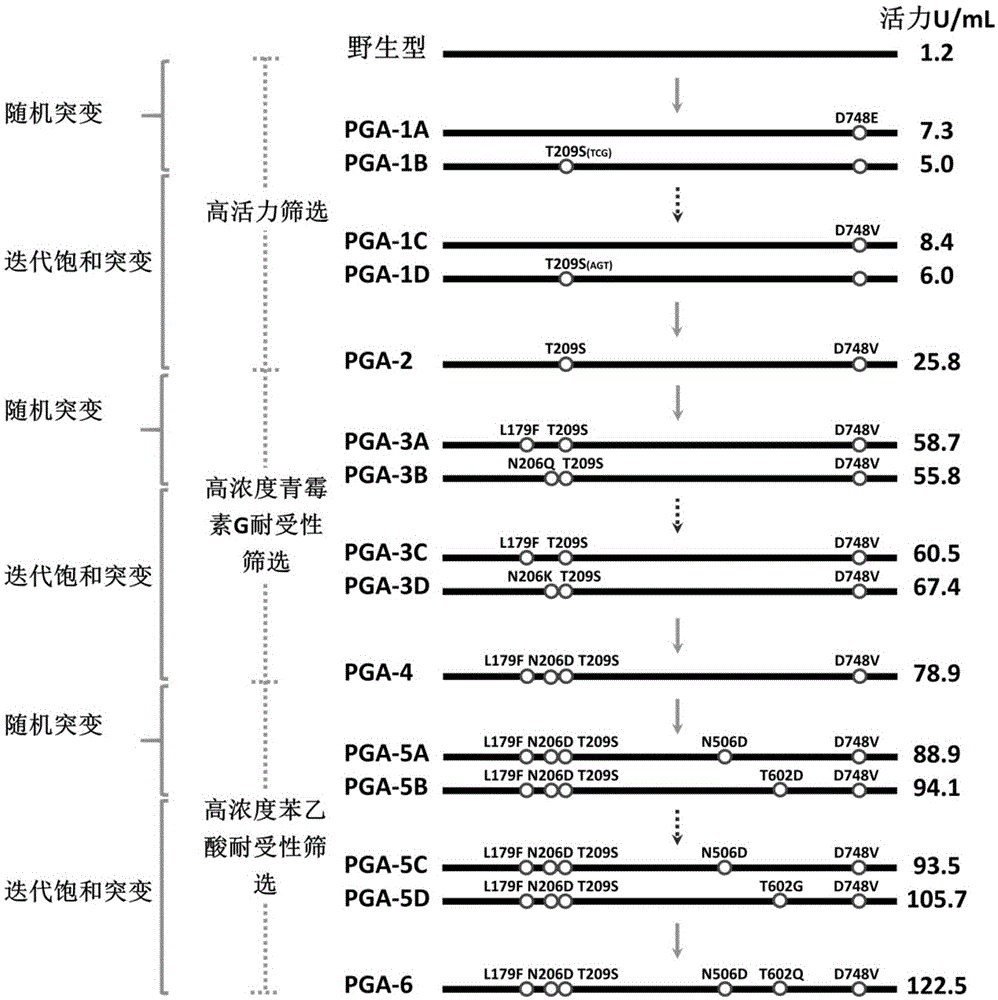
[0084] Embodiment 2: the preparation of PGA mutant
[0085] 2-1 Preparation of highly active PGA mutants
[0086] 2-1-1 Construction of PGA Random Mutant Library
[0087] In order to improve the activity of wild-type PGA to penicillin G, the inventors used PGA-WT as a template, wherein the primers were T7 universal primers (SEQ ID NO: 23 and 24), constructed a random mutant library by error-prone PCR, and passed Adjusting Mg in error-prone PCR reaction system 2+ and Mn 2+ concentration and dCTP and dTTP oligonucleotide concentrations, the base mismatch rate of the mutant library is only 2 / 1000, that is, it is guaranteed that only 1 to 2 amino acids are mutated in a mutant. The specific process of constructing the mutant library is as follows.
[0088] Error-prone PCR reaction system:
[0089]
[0090] The error-prone PCR reaction conditions are: 95°C pre-denaturation for 5 minutes; then 94°C denaturation for 30 seconds, 56°C annealing for 1 minute, 72°C for 1.5 minutes...
Embodiment 3
[0154] Application of embodiment 3 mutant PGA-6 immobilized enzyme
[0155] 3-1 Wild-type PGA and PGA-6 immobilized enzyme cleave penicillin G to prepare 6-APA experiment
[0156] (1) Cleavage reaction
[0157] The above-mentioned wild-type PGA and PGA-6 immobilized enzymes were respectively placed in penicillin G solutions of certain concentrations (8%, 15%, 20%, 25% and 30%) with the same enzyme amount (8000U), at 25°C , react under the condition of pH8.00, in the reaction process, continuously drip 3mol / L of concentrated ammonia water to neutralize the acid produced in the reaction process, make the pH constant at 8.00, and record the amount of ammonia water added, when the reaction reaches the end point (judgment end point The best method is: automatically stop adding ammonia water, the pH value remains unchanged for more than 3 minutes), and record the total reaction time. Filter the above lysate, rinse the immobilized enzyme with sterile deionized water 4-5 times repea...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Vitality | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com