Novel cucumber SNP molecular marker
A molecular marker, cucumber technology, applied in the biological field, can solve the problems of complex, long genetic distance between disease resistance genes and molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
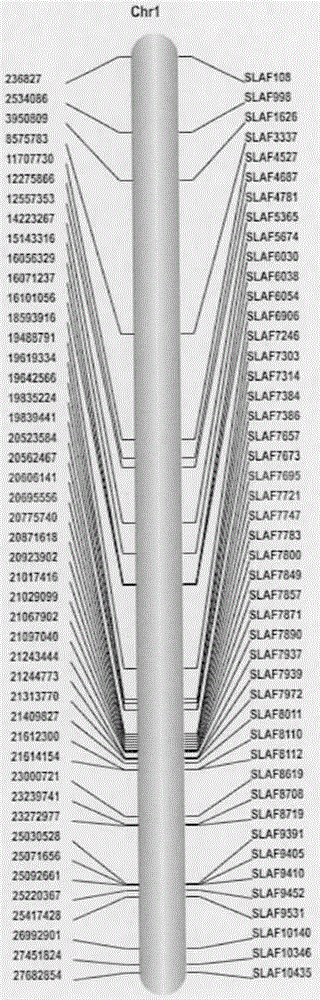
[0024] The young leaves of the material at the stage of two leaves and one heart were taken, and the genomic DNA was extracted by the improved SDS method, diluted to 60 ng / μL, and the concentration was detected by 1% agarose electrophoresis. The enzyme digestion prediction software SLAF_Predict system was used to analyze the cucumber genome, and the enzyme digestion scheme, gel cutting range and sequencing volume were determined according to its GC content, repeat sequence and gene characteristics, so as to ensure the density and uniformity of molecular markers.
[0025]Take 500ng of genomic DNA, add 0.6U Mse I (NEB, Hitchin, Herts, UK), T4 DNA ligase (NEB), ATP (NEB) and Mse I linker (NEB), react at 37°C for 15h, anneal at 65°C 1h, then add restriction endonucleases Hae III and Bfa I and react at 37°C for 3h. After the reaction, the digested product was purified with Quick Spin column (Qiagen), and redissolved with 33 μL EB. Use the recovered digested product as a template f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com