Photobacterium leiognathi luciferase, gene coding photobacterium leiognathi luciferase, and applications of photobacterium leiognathi luciferase
A technology of luciferase gene and photobacillus sp. is applied in application, genetic engineering, plant genetic improvement and other directions, which can solve the problems of high cost and less preparation of enzyme solution.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Cloning and expression of luciferase gene LuxAB in Escherichia coli RosettaDE3
[0022] PCR-amplified the LuxAB gene encoding luciferase in Photobacterium chinensis, and digested with restriction endonucleases BamHI and XhoI, and then recovered using an agarose gel DNA recovery kit (BPI); The final plasmid pET-28a(+) was subjected to a ligation reaction, and then the ligated product was introduced into the expression host Escherichia coli RosettaDE3. After the sequencing was correct, the expression was induced with 0.5mMIPTG for 10h, and purified by Ni Sepharose TM 6Fast Flow filler Obtain active recombinant luciferase protein.
Embodiment 2
[0023] Example 2 Detection and verification of FMN:NADH activity in Escherichia coli RosettaDE3
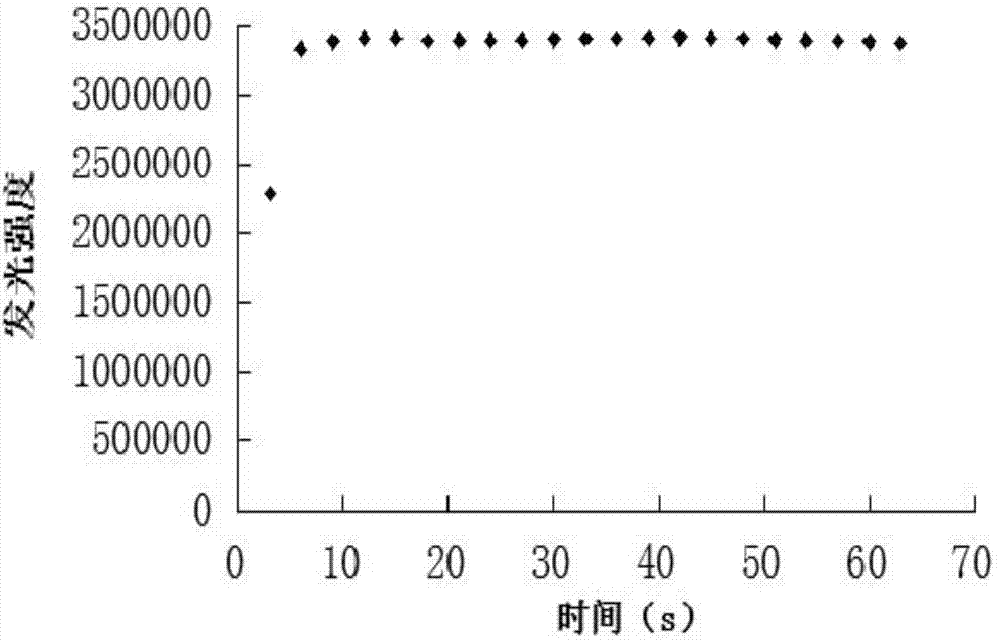
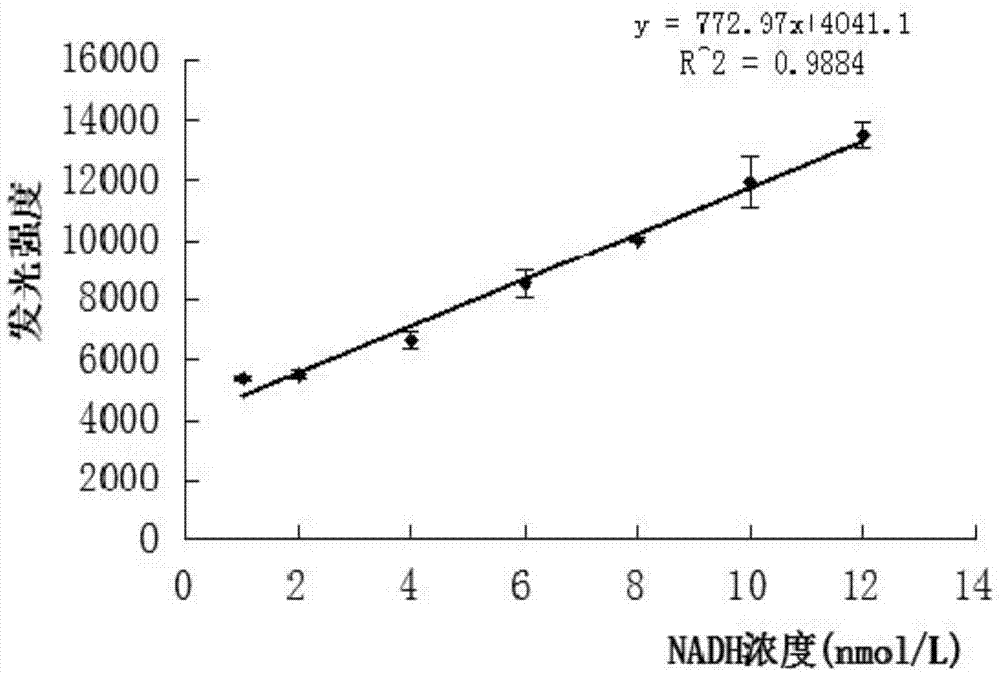
[0024] The crude enzyme solution extracted from Escherichia coli RosettaDE3 by sonication and centrifugation, 1ml of the enzyme solution was taken, and the substrates FMN-Na 0.5μL (10mM) and NADH 300μL (0.14mM) were quickly added at one time, and the absorbance value was measured. The absorbance value is measured by scanning with a UV fluorescence spectrophotometer. The detection conditions: set the wavelength to 340nm. After adding the solution or reagent, perform a time scan. The decrease in the absorbance value within 60s can reflect the activity of FMN:NADH oxidoreductase.
[0025] FMN:NADH oxidoreductase activity assay results in Escherichia coli rosettaDE3 are shown in Table 1.
[0026] Table 1
[0027]
[0028] In Table 1, the FMN reductase activity of Escherichia coli without the luciferase gene was almost equal to that of the recombinant Escherichia coli with the luci...
Embodiment 3
[0030] Example 3 Preparation of recombinant Escherichia coli RosettaDE3 in vitro dual enzyme crude enzyme preparation
[0031] At 24°C, 150r / min, induce 10h of recombinant Escherichia coli RosettaDE3300mL, collect the bacteria by centrifugation, and reconstitute the bacteria with 0.01mol / L PBS (pH 7.0, containing 10mmol / L EDTA, 1mmol / L DTT, the same below). Dissolving, ultrasonic crushing in an ice bath, ultrasonic time 15s, interval 15s, crushing 60 times, power 300W; 4°C refrigerated centrifugation (9000r / min, 30min), supernatant was salted out with 40% to 80% solid ammonium sulfate, centrifuged , the precipitate was redissolved in 30 mL of PBS, and dialyzed in PBS at 4°C for 24 hours to obtain the crude enzyme solution for the experiment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com