Design, synthesis and use of RNA molecule for high-efficiency genome editing
A base and base number technology, applied in the field of genetic engineering and nucleic acid detection, can solve the problems of application limitations and low cutting efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
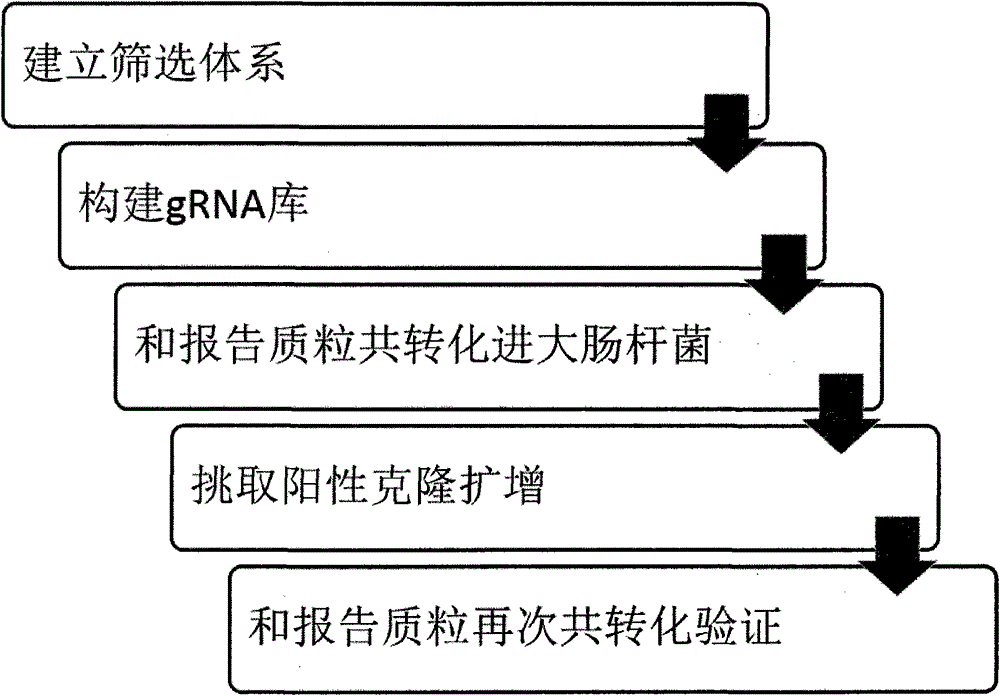
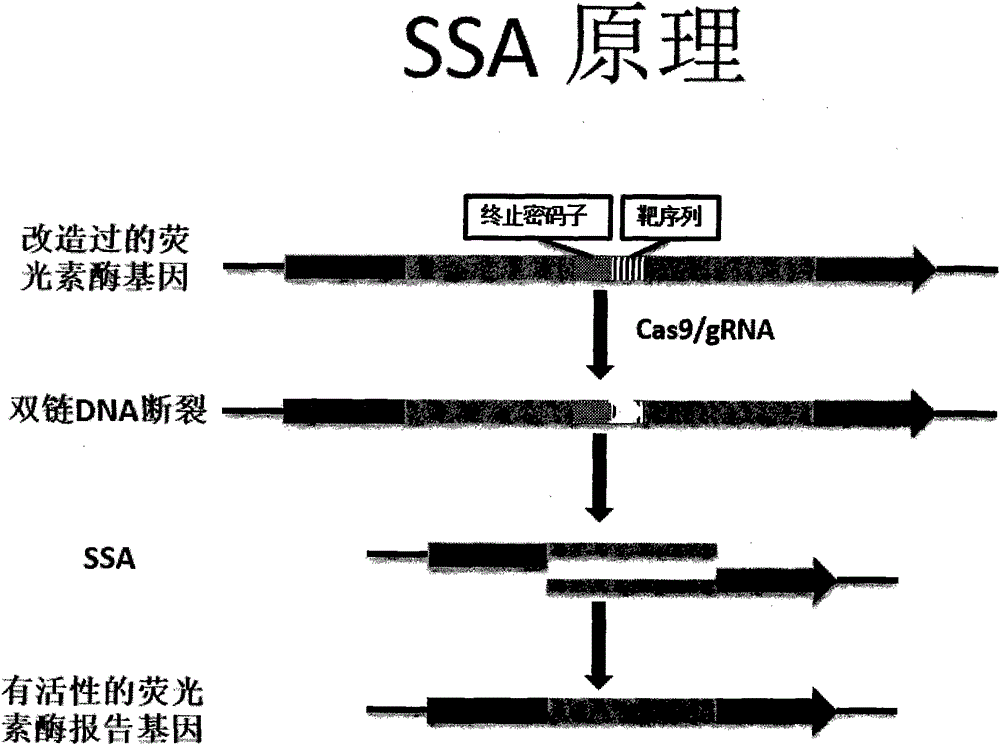
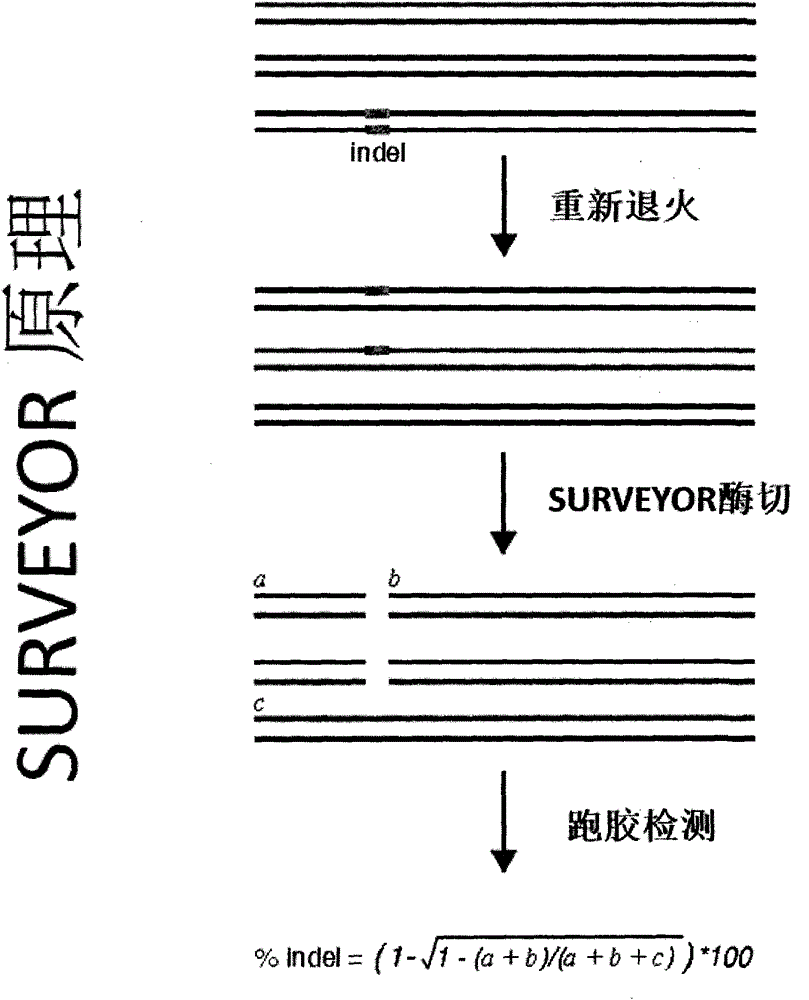
[0042] The principle of embodiment 1 is as follows figure 2 with image 3 As shown, the experimental results are as Figure 18 shown. First, we constructed the gRNA recognition sequence into the SSA vector, and then constructed the gRNA expression vector on the eukaryotic expression vector; secondly, we co-transfected the gRNA expression vector, cas9 expression vector, and SSA expression vector into HEK293T cells for 24 hours The expression level of the luciferase gene was then checked. At the same time, we co-transfected the gRNA expression vector and the cas9 expression vector into HEK293T cells. After 48 hours, we extracted the genomic DNA and performed the SURVEYOR experiment to reflect the gRNA / cas9 activity. from Figure 18 It can be seen that the activity of gRNA1 is about 50% higher than that of gRNA.
Embodiment 2
[0043] The experimental process of Example 2 is similar to that of Example 1, except that the sequence of gRNA1 is replaced by gRNA2. See the experimental results Figure 19 . Depend on Figure 19 It can be seen that the activity of gRNA2 is increased by about 30% relative to the activity of gRNA.
Embodiment 3
[0044] The experimental process of Example 3 is similar to that of Example 1, except that the sequence of gRNA1 is replaced by gRNA3. See the experimental results Figure 20 . Depend on Figure 20 It can be seen that the activity of gRNA3 is about 30% higher than that of gRNA.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com