L-threonine aldolase from enterobacter cloacae and application thereof
A technology of threonine aldolase and Enterobacter cloacae, applied in the field of chemical engineering, can solve problems such as high cost, serious environmental pollution, and complicated process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
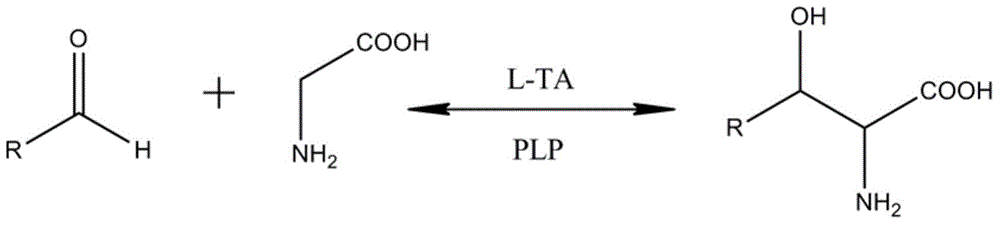
[0042] Example 1 Isolation of the nucleotide sequence of L-threonine aldolase from Enterobacter cloacae
[0043] The total DNA was extracted from Enterobacter cloacae cells cultured for 36 hours according to the method provided in the "Molecular Cloning Experiment Guide", and 7 μg was used as a template for polymerase chain reaction. According to the published L-threonine aldolase sequence design, PCR amplification was carried out on a T-Gradient PCR instrument (Biometra Company) using the above-mentioned extracted DNA as a template. The primers, components and amplification conditions used in the reaction were as follows:
[0044] Primer 1: 5'-ATGATTGATTTACGCAGTGATACCG-3'
[0045] Primer 2: 5'-TTAACGCTGTAAAAACGCCTGCCAG-3'
[0046]
[0047] Amplification conditions: Denaturation at 94°C for 3min, followed by 30 cycles of 94°C for 1min, 58°C for 1min, 72°C for 1min, and finally 72°C for 10min. The results of agarose gel electrophoresis showed that a fragment with a size of...
Embodiment 2
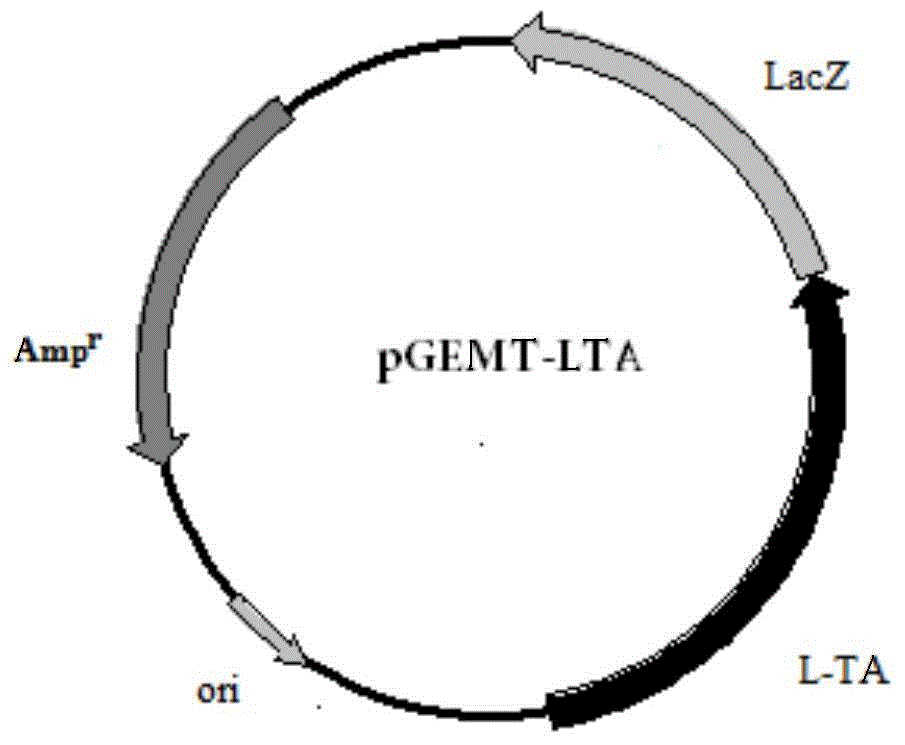
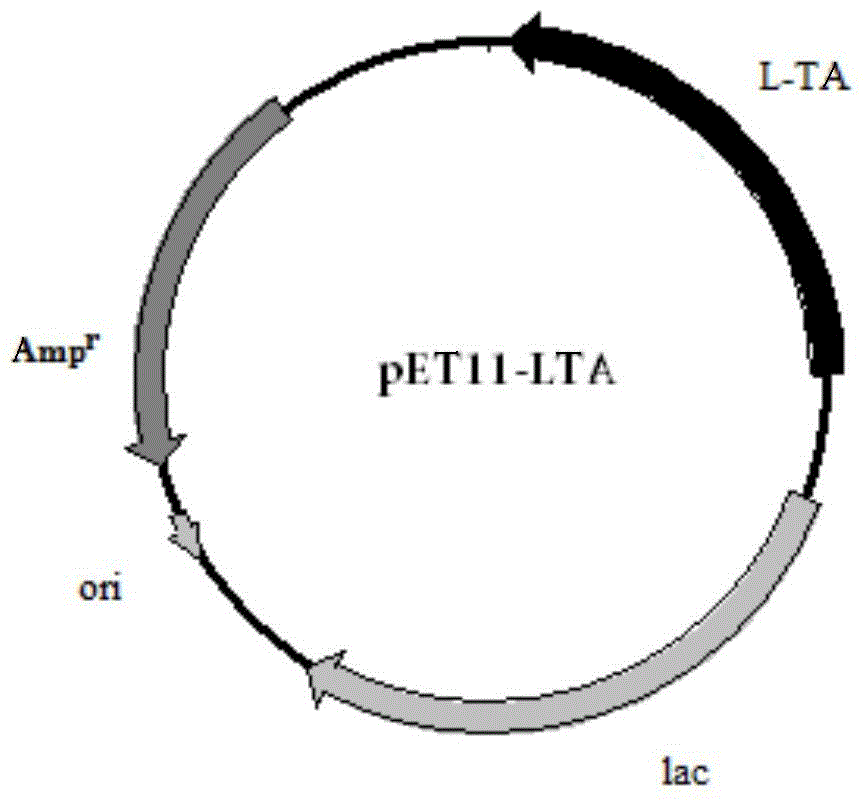
[0048] Embodiment 2: Construction of Escherichia coli recombinant expression vector
[0049] According to the coding region sequence shown in SEQ ID NO:1, a pair of gene-specific amplification primers is designed to separate its potential open reading frame sequence:
[0050] Primer 3: 5'-GGCTCTAGA ATGATTGATTTACGCAGTGATACCG-3';
[0051] Primer 4: 5'-GCCGGATCC TTAACGCTGTAAAAACGCCTGCCAG-3';
[0052] The 5' ends of these two primers contain XbaI and BamHI restriction sites respectively. The amplification conditions and reaction components used are the same as above, and the sequencing result of the amplified product shows that it is consistent with the sequence shown in SEQ ID NO:1. Then, 50 μl of the PCR product and 1 μl of pET11a (Invitrogen) were subjected to double enzyme digestion, and the large fragment was recovered and ligated with T4 ligase overnight in a 4-degree refrigerator. The ligation product was transformed into Escherichia coli DH5α, and positive clones were s...
Embodiment 3
[0053] Example 3: Preparation of L-threonine aldolase crude enzyme
[0054] Inoculate a single colony of the identified positive transformant in SOC medium containing Amp (100 μg / ml), culture it with shaking at 37°C overnight, and the bacterial concentration reaches OD 600 When =1, inoculate 1% in LB medium containing Amp (100μg / ml), and continue to culture at 37°C until OD 600 At 1.2-1.5, the cells were harvested by centrifugation. The cells were resuspended in 0.1M phosphate buffer solution, ultrasonically disrupted, and then centrifuged at 20,000 rpm for 1 hour. The supernatant was harvested as the crude enzyme of L-threonine aldolase, aliquoted, and stored at -20°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com