Anti-rice blast gene Pi9 specific CAPS marker Pi9caps and application thereof
A rice blast resistance gene and rice blast technology, applied in the field of crop molecular genetics and breeding, can solve problems such as low selection efficiency, difficulty in rice blast resistance genes, and high chance of polymorphism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073]Example 1, Primer Design and Detection of Rice Blast Resistance Gene Pi9 Gene-Specific CAPS Marker Pi9caps
[0074] 1. Analysis of Pi9 gene sequence-specific SNP sites
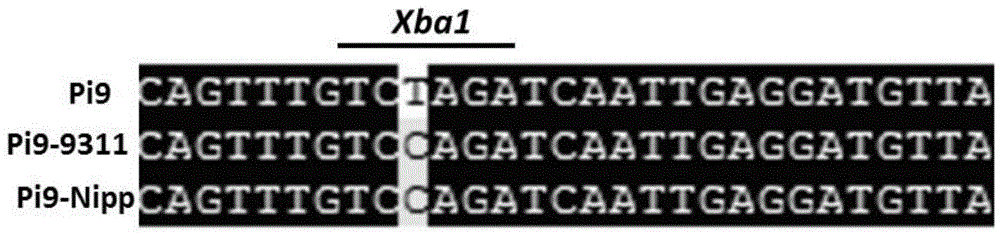
[0075] The published Pi9 gene and the sequenced genome sequences of the corresponding regions of rice varieties 93-11 and Nipponbare were downloaded from the public database, and the sequences were compared against the Pi9 locus to screen for Pi9-specific genes that could be differentiated from those genes. SNPs for locus alleles. Wherein the T base located at 7080bp after the start codon of the Pi9 gene (that is, the nucleotide sequence of 75-1-127BAC12.1 in rice chromosome 6 from the 5' end to the 40804th position) is at 93-11 and Nipponbare The alleles in are all C, and this SNP is just located in the recognition sequence of restriction endonuclease XbaI (recognition sequence is 5'-TCTAGA-3') (such as figure 1 as shown, figure 1 Among them, Pi9 is the rice blast resistance gene Pi9, Pi9-9311 is the...
Embodiment 2
[0097] Example 2, Verification and Application of Rice Blast Resistance Gene Pi9-specific CAPS Marker
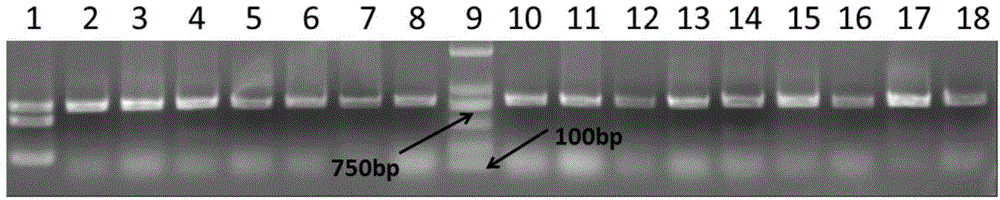
[0098] 1. Using the rice variety 93-11 without the blast resistance gene Pi9 as the female parent, and the rice variety 75-1-127 containing the blast resistance gene Pi9 as the male parent, the F1 generation was obtained, and the F1 generation was obtained by selfing For the F2 population, extract the genomic DNA of individual plants numbered 4-24, 93-11 and 75-1-127 in the F2 population.
[0099] Two, the genomic DNA obtained in step 1 is used as a template respectively, and the primers F and R of Example 1 are used as primers to carry out PCR amplification to obtain a PCR amplification product with a size of 802bp. This PCR amplification product is the Pi9caps marker , the marker is the 75-1-127BAC12.1 sequence from the 5' end to the 41006th position of the 75-1-127BAC12.1 sequence in the No. 6 chromosome of each rice, and then use the restriction endonuclease XbaI to dige...
Embodiment 3
[0106] Example 3. Verification and application of rice blast resistance gene Pi9-specific CAPS marker in rice transfected with Pi9 gene coding region
[0107] 1. Using the rice variety TP309 without the rice blast resistance gene Pi9 as a transgenic acceptor, overexpressing the CDS region of the rice blast resistance gene Pi9 in TP309 to prepare rice transgenic for the coding region of the Pi9 gene.
[0108] Two, the genomic DNA of the rice and TP309 of the transgenic Pi9 gene coding region that extraction step 1 obtains, use it as template respectively, take the primer F and R of embodiment 1 as primer, carry out PCR amplification, obtain each PCR amplification product, Restriction endonuclease XbaI was used to digest each PCR amplification product to obtain each digested product, and each digested product was detected by electrophoresis on an agarose gel.
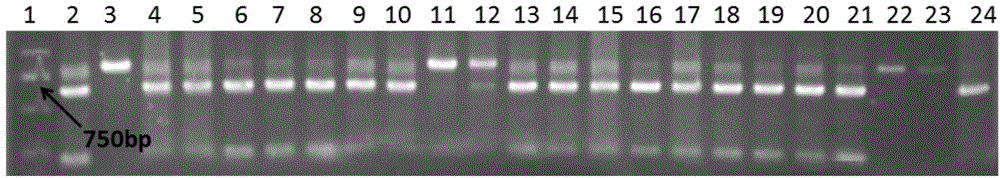
[0109] The result is as Figure 4 shown.
[0110] Figure 4 Among them, lane 1 is the DNA ladder, and lanes 2 and 3 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com