Method for detecting non-coding RNA transcription level based on hybridization connection method
A transcription level, non-coding technology, applied in the field of accurate measurement of non-coding RNA transcription levels, can solve the problems of unstable reverse transcription non-coding RNA efficiency and non-reproducibility, and achieve superior detection environment, reproducibility and accuracy The effect of increasing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
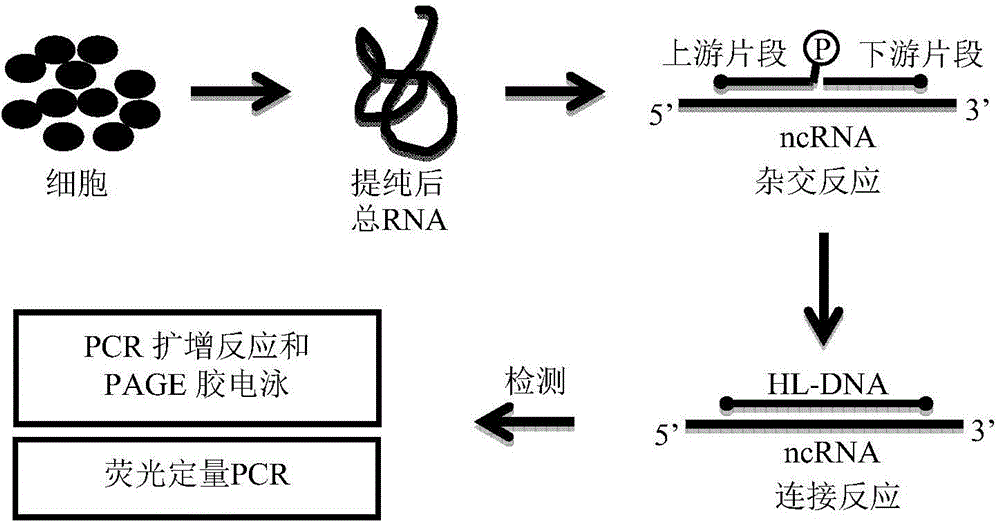
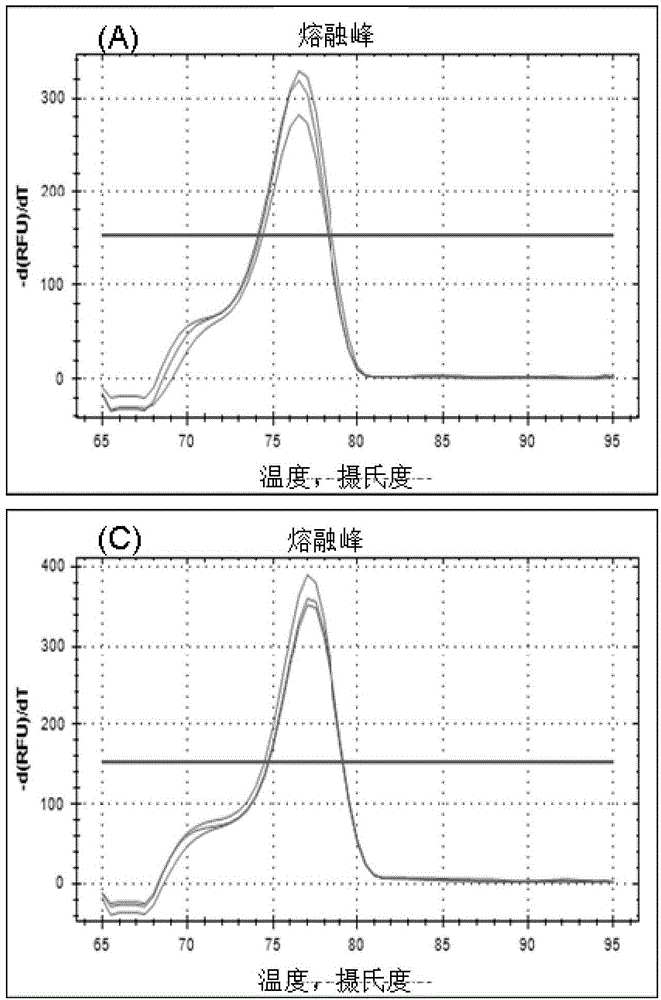
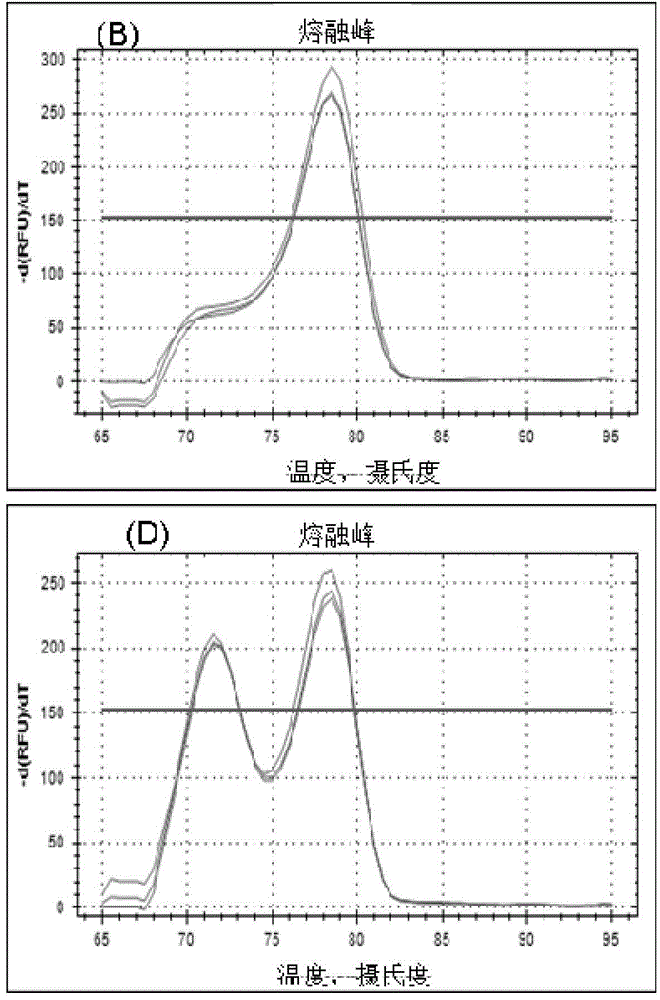
[0055] a kind of like figure 1 The method for detecting non-coding RNA transcript levels based on the hybridization ligation method of the present invention as shown, comprises the following steps:
[0056] 1. Cell culture in vitro
[0057] HeLa cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai), and the cells were placed in DMEM medium (cell culture medium) containing 10% FBS (fetal bovine serum), 100 U / ml penicillin and 0.1 mg / ml streptomycin. At 37°C, 5% CO 2 cultured in a humidified incubator.
[0058] 2. Extraction of total RNA
[0059] The steps of RNA extraction by Trizol method are as follows: ①Take out the cells from the incubator, suck off the old culture medium, wash the cells quickly with 1×PBS preheated at 37°C, suck out the residual PBS, and then add 3~5mL of 37°C preheated 1 x PBS. ② Use a cell scraper to quickly scrape the cells, collect them in a 10mL centrifuge tube, put them in a refrigerated centrifuge (pre-cooling is...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com