Nitrile hydratase gene, encoded enzyme, vector, engineering bacterium as well as application of engineering bacterium to preparation of amide compound
A nitrile hydratase and gene-encoded technology, applied in genetic engineering, using vectors to introduce foreign genetic material, applications, etc., can solve problems such as low activity and limited applications, and achieve high-efficiency transformation effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
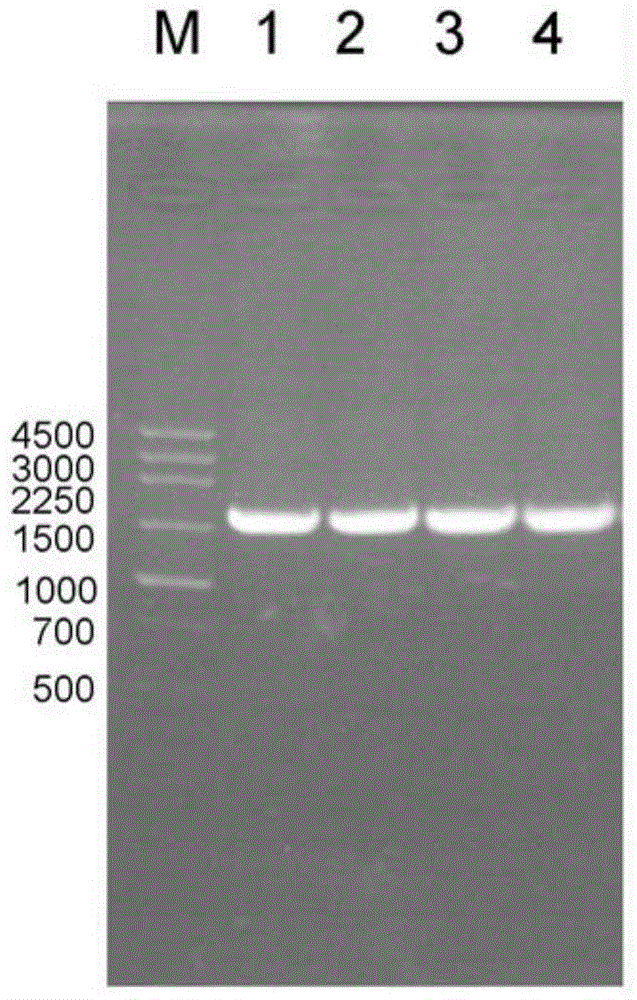
[0024] Example 1: Cloning of full-length nitrile hydratase gene
[0025] The genome of Sulphitobacter sp. E-36ATCC BAA-1142 was extracted using a bacterial genome extraction kit. Design primers NH_E36-Up and NH_E36-Down according to the nucleotide sequence (such as SEQ ID NO.1) of hypothetical nitrile hydratase in the Bacillus bisulfite E-36 genome again, take Bacillus bisulfite E-36 genome as template, PCR amplifies Amplify the full-length nitrile hydratase gene, such as figure 1 shown. The PCR product is recovered using a DNA gel recovery and purification kit, and the obtained full-length nitrile hydratase gene is composed of the alpha subunit shown in SEQ ID NO.2 (the encoded amino acid sequence is shown in SEQ ID NO.5), SEQ ID NO The β subunit shown in .3 (the encoded amino acid sequence is shown in SEQ ID NO.6) and the activation element shown in SEQ ID NO.4 (the encoded amino acid sequence is shown in SEQ ID NO.7) consists of SEQ ID NO . The α subunit shown in 2, the ...
Embodiment 2
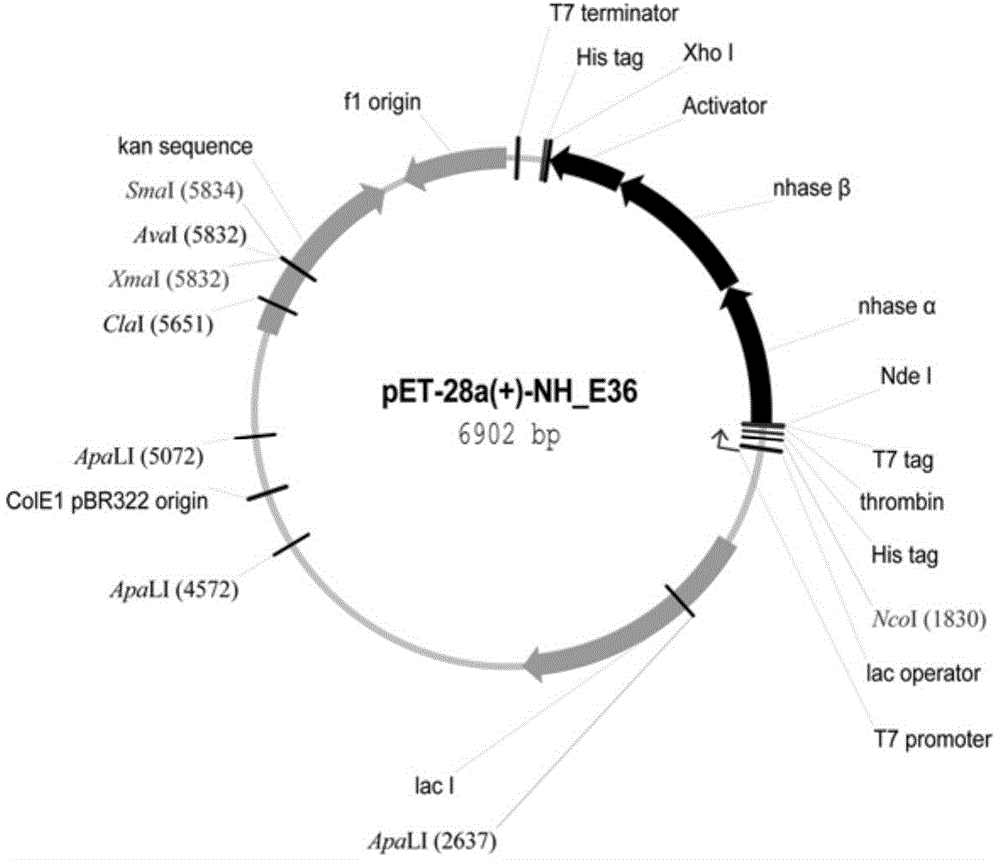
[0036] Embodiment 2: Construction and identification of genetic engineering strain E.coli pET28a(+)-NH_E36
[0037] In order to achieve efficient functional expression of the nitrile hydratase gene derived from the Bacillus bisulfite E-36 genome in Escherichia coli BL21(DE3) cells, pET28a(+) containing T7 promoter was selected as the expression vector. The vector pET28a(+) and the full-length nitrile hydratase gene were double-digested with Nell and Xho I, and the digested product was recovered using a DNA gel recovery kit.
[0038] Double enzyme digestion system and reaction conditions:
[0039]
[0040] Incubate at 37°C for 5h.
[0041] Nucleic acid electrophoresis was used to preliminarily determine the concentration of the two, and the gene / plasmid (mol / mol, 2:1) was mixed, and T4 DNA ligase was added to connect overnight at 16°C to obtain the recombinant plasmid pET28a(+)-NH_E36 (schematic diagram as shown in figure 2 shown). Then, the recombinant plasmid was trans...
Embodiment 3
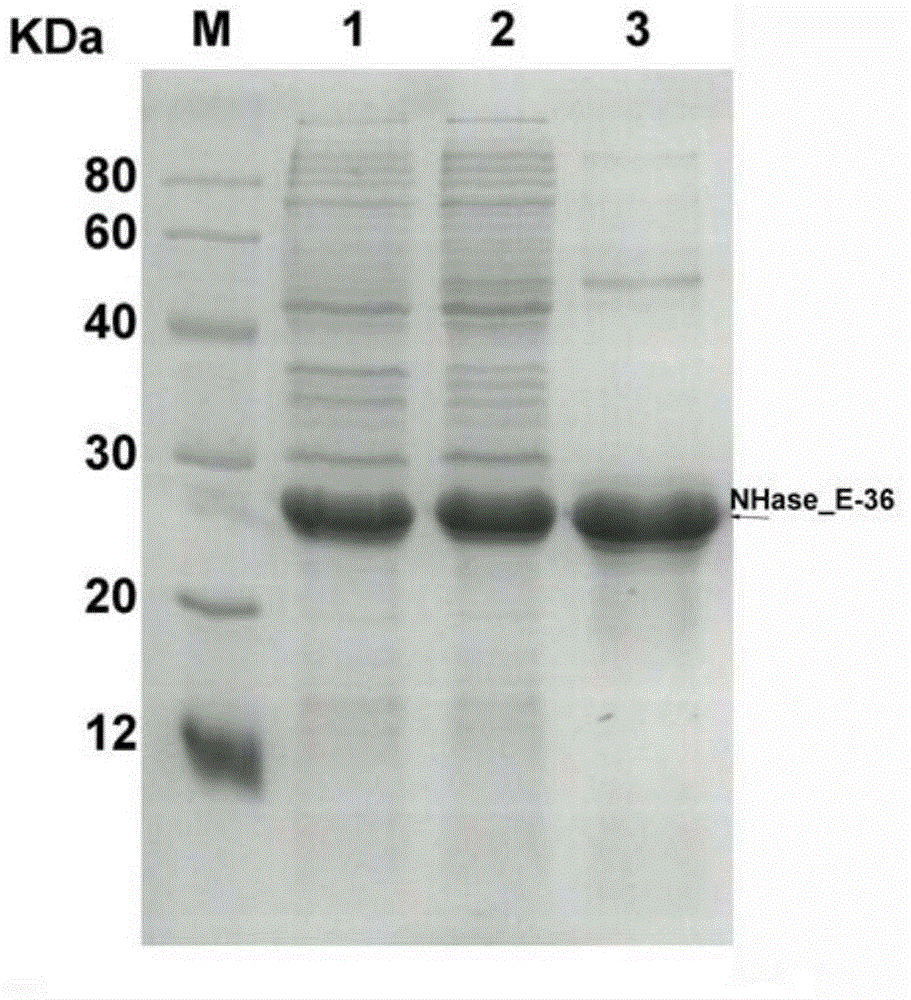
[0042] Example 3: Expression and purification of nitrile hydratase of genetic engineering strain E.coli pET28a(+)-NH_E36
[0043] The genetically engineered bacteria E.coli pET28a(+)-NH_E36 glycerol-preserved strain was inoculated into 5 mL of LB liquid medium containing 50 μg / ml Kan, and cultured at 37° C. with shaking at 200 rpm for 10 to 14 hours. Transfer 2 mL of the culture solution to 100 mL of fresh LB liquid medium containing 50 μg / ml Kan, and culture at 37 °C with shaking at 200 rpm until the cell density (OD 600 ) reaches about 0.8, add IPTG to a final concentration of 0.1 mM, and induce at 18-37°C for 12-18 hours. Cells were collected by centrifugation at 5000-10000×g for 5-10 min, washed twice with phosphate buffer (50-200 mM, pH 5.0-8.0), and resuspended in phosphate buffer. In an ice bath, use an ultrasonic breaker to break the cells, centrifuge at 5000-10000×g for 5-30 minutes, collect the supernatant of the broken cells, and pass the supernatant of the broken ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com