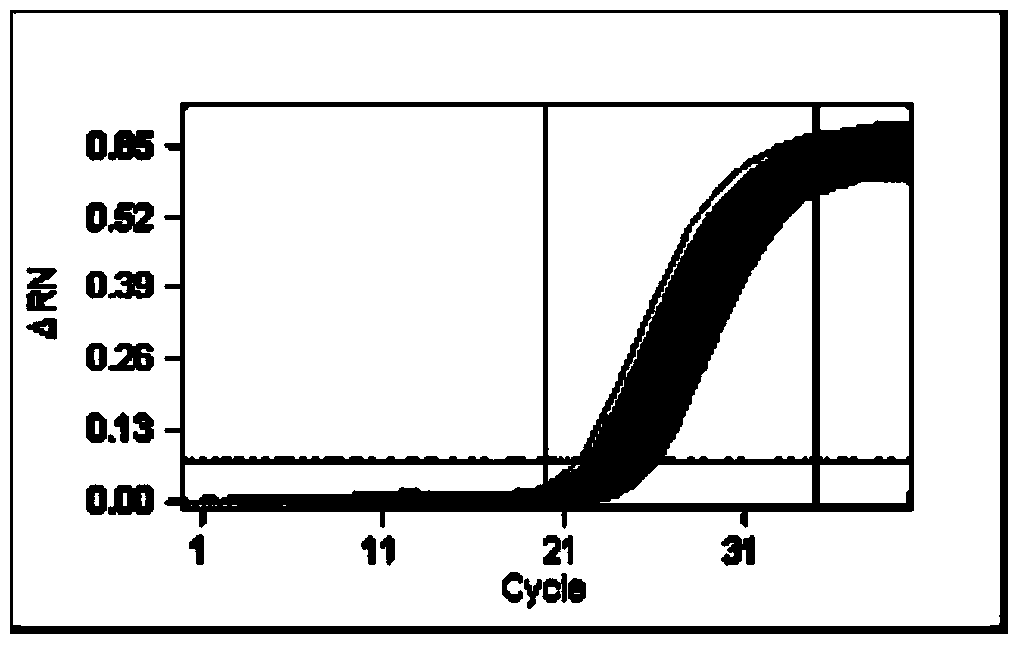
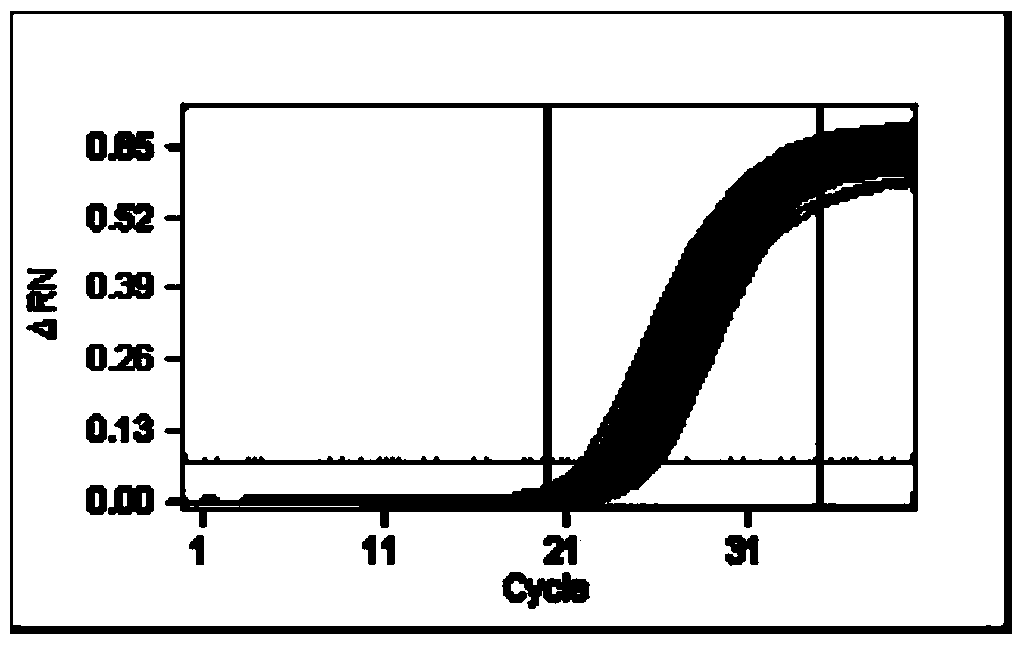
Method for identifying homozygotic type or heterozygous type NK603 on basis of digital PCR (polymerase chain reaction)
A heterozygous and homozygous technology, applied in the field of identification of homozygous or heterozygous NK603 based on digital PCR, can solve the problems of time-consuming, large error, and difficult result judgment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0083] The following is a detailed description in conjunction with the drawings and embodiments:
[0084] 1. Using CTAB method to extract genomic DNA of genetically modified corn NK603
[0085] 1) Frozen the mortar and pestle at -20°C for more than 10 hours, and heat 1.5×CTAB extract at 100°C, and preheat 10% CTAB to 56°C; ground 8.0 g of genetically modified corn NK603 fresh with liquid nitrogen Leaf, transfer the powder to a 50ml centrifuge tube; add 20ml of preheated 1.5×CTAB extract to the centrifuge tube, stir evenly with a glass rod, put it in a 56℃ water bath for 40 minutes, gently invert it from time to time ; After a water bath, cool to room temperature; add 20 ml of chloroform / isoamyl alcohol (24:1), cover, invert evenly until it becomes turbid, centrifuge at 4000 r / min for 20 min at room temperature; transfer the supernatant to a new 50 In a ml centrifuge tube, add 1 / 10 volume of 10% CTAB solution preheated at 56°C, then add an equal volume of chloroform / isoamyl al...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com