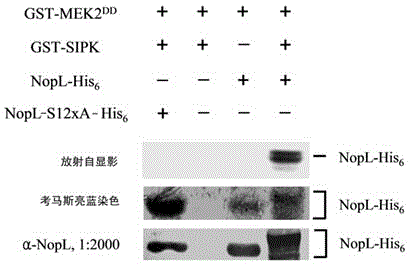
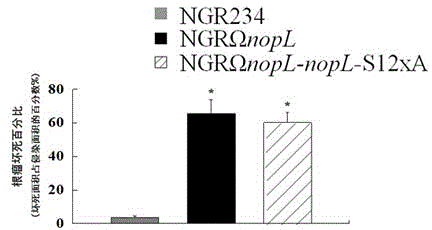
Application of phosphorylated bacterioprotein NopL
A phosphorylation, protein technology, applied in the field of molecular biology, can solve problems such as unclear mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Construction of recombinant plasmid pBS- nopL -S12xA
[0047] 1. The present embodiment utilizes artificially synthesized DNA fragments and PCR to carry out site-directed mutagenesis to construct a recombinant plasmid pBS- nopL -S12xA. First synthetic mutant recombinant gene nopL -S12xA, the recombinant protein NopL-S12xA contains 12 mutated phosphorylation sites, all of which are point mutations from serine (Serine) to alanine (Alanine). The 12 mutation sites are: S7A / S12A / S17A / S36A / S73A / S89A / S139A / S148A / S187A / S198A / S240A / S245A.
[0048] 2. Among the above 12 mutation sites, the 5 point mutations S7A / S12A / S17A / S36A / S89A at the N-terminal were mutated by the method of artificially synthesizing nucleotide fragments. The artificially synthesized DNA fragment sequence (314 bp) is as follows: SEQ ID NO: 3, both ends contain Eco RV- Asc I restriction site for further molecular cloning.
[0049] 3. The remaining 7 point mutations were completed by PCR
...
Embodiment 2
[0089] Example 2 Construction of recombinant expression vector pPROEX- nopL -S12xA
[0090] The purpose of this example is to convert gene fragments nopL - S12xA was cloned into the pPROEX expression vector.
[0091] First design primers NopL-S12xA-F and NopL-S12xA-R, and introduce them at both ends of the primers Ehe I and Bam HI restriction site (underlined) to facilitate its insertion into the pPROEX vector.
[0092] The primer sequences are as follows:
[0093] NopL-S12xA-F: (as shown in SEQ ID NO: 16)
[0094] 5’-CGG GGCGCC ATGGATATCAATTCAACCGC-3';
[0095] NopL-S12xA-R: (as shown in SEQ ID NO: 17)
[0096] 5'- GCGGATCC TCAAATGTCAAAATCCACCG-3'.
[0097] Take pBS- nopL -S12xA is used as the template, and the above primers are used to carry out the PCR amplification reaction of exogenous fragments. The reaction system (total volume 50 μL) is as follows:
[0098] Template (50 ng) 1 μL
[0099] dNTP Mix (2.5 mM) 4 μL
[0100] Primer F (10 μM) 1 μL
[01...
Embodiment 3
[0110] Example 3 Induction, expression and purification of recombinant protein NopL-S12xA
[0111] 1. Induction and expression of recombinant protein NopL-S12xA
[0112] The recombinant engineered bacteria BL21 (DE3- nopL- S12xA) were streaked into LB solid medium containing ampicillin (50 μg / mL) and incubated at 37°C for 12-16 h.
[0113] A single colony was randomly selected and inoculated into 3 mL of LB liquid medium containing ampicillin (50 μg / mL), and cultured with shaking at 37 °C and 220 rpm for 12 h.
[0114] The cultured bacterial solution was inoculated into 300 mL of LB liquid medium containing ampicillin (50 μg / mL) at a ratio of 1:100, and incubated at 37°C with shaking to OD. 600 = 0.6 (about 3h), add IPTG to a final concentration of 1mM, and continue to shake at 37°C and 220rpm for 20h.
[0115] 2. Purification and recovery of recombinant protein NopL-S12xA
[0116] The above-mentioned bacterial solution induced by IPTG was centrifuged at 5500 g for 5 mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com