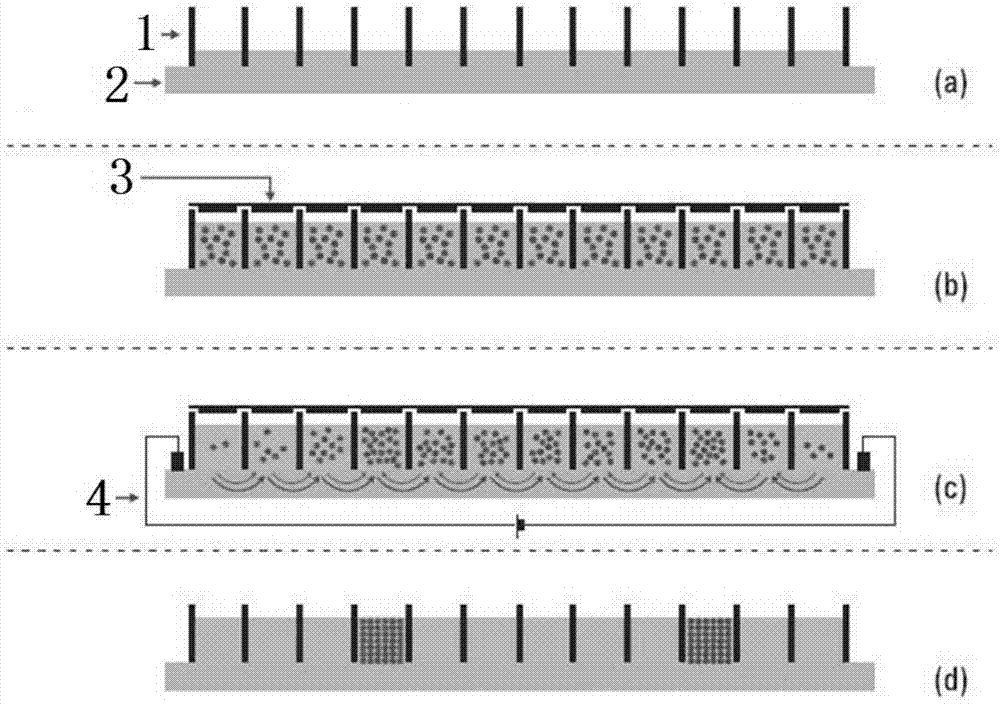
Polypeptide mixture isoelectric focusing separation method for proteomics analysis
A technology of proteomics and isoelectric focusing, applied in the field of proteomics, can solve the problems of cumbersome operation, affecting the detection of low-abundance proteins, and poor reproducibility of experimental results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The following reagents, acetonitrile, water, and formic acid, were all chromatographically pure and were purchased from Sigma-Aldrich Reagent Company; mass spectrometry-grade trypsin was purchased from Promega Company; dry strips and sample cups were purchased from Bio-Rad Company; the mass spectrometer was a linear ion Trap-Electrostatic Field Orbitrap Combined Mass Spectrometer (Thermo LTQ-Orbitrap Elite).
[0028] (1) After ultrasonic crushing of buffalo sperm, add lysate (7M urea, 2% CHAPS, 0.5% DTT and 0.5% PMSF) to digest and extract total protein, centrifuge to remove cell debris and other impurities, and discard the lower layer of cell debris;
[0029] (2) Take 500 μL of the supernatant extracted above, add 1.5 mL of pre-cooled acetone to precipitate overnight, centrifuge at 12,000 rpm for 1 h, discard the supernatant, and wash the precipitate with 100 μL of 40 mM ammonium bicarbonate (NH 4 HCO 3 ) reconstitution;
[0030] (3) Add 10 μL 40 mM dithiothreitol (D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com