Method of knocking out spinal muscular atrophy SMN genes and cell model
A spinal muscular atrophy and gene knockout technology, applied in the field of genetic engineering, can solve problems that have not been seen yet, achieve good stability and reliability, low cost, and simple screening methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Knockout of SMN gene
[0034] 1. Design and construction of SMN gene ZFN plasmid knockout
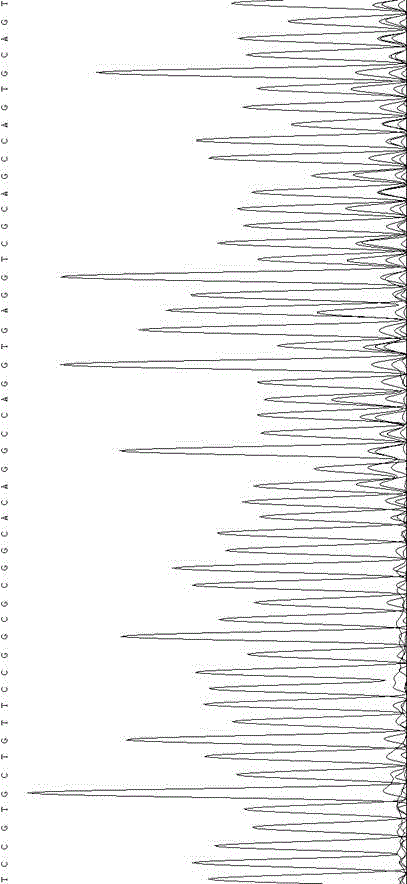
[0035] According to the principle of ZFN technology, the DNA targeting recognition site is designed. The site is designed at the first exon shared by SMN1.SMN2 transcripts. After a lot of repeated tests, the determined target sequence information is:
[0036] TCCGTGCTGTTCCGGCGC ggcac AGGCCAGGTGAGGTCGCA (SEQ ID NO. 1). The underlined part at both ends is the ZFN binding region, and the lowercase letter in the middle is the ZFN cutting region. The plasmid construction was completed and identified by Sigma, USA. The specific information of its ZFN plasmid is as follows: figure 1 shown.
[0037]The specific construction method of the artificial zinc finger nuclease expression vector, the expression vector backbone is pAVX (Sigma), the vector contains pUC oir to promote the replication of the plasmid in prokaryotic cells, and has a kanamycin resistance gene; at...
Embodiment 2
[0073] Example 2 SMN gene knockout cell line mRNA expression identification
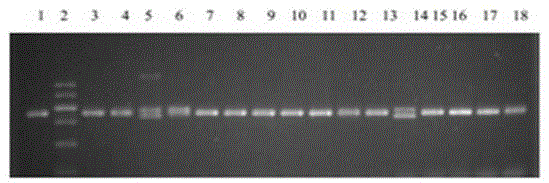
[0074] For the identification of cells with effective SMN knockout, extract RNA. For the method, refer to the instruction manual of the Trizol kit. After the total RNA is extracted, it is digested with DNase without RNAase activity to remove traces of genomic DNA. The RNA content is measured by UV spectrophotometer and confirmed by electrophoresis. RNA quality; cDNA was synthesized according to the instructions of the reverse transcription kit from Tiangen Company. Different primers were designed for (C → T) in the seventh exon of SMN1 and SMN2, and RT-PCR was carried out using different transcripts of SMN1 and SMN2 as templates to preliminarily determine the deletion of SMN1 and SMN2 according to the semi-quantitative situation.
[0075] However, we found that base deletions and base insertions in exons did not affect the stability of RNA transcribed by SMN1 and SMN2. Based on this conclusion, we...
Embodiment 3
[0096] Example 3 Western blot detection of SMN protein expression
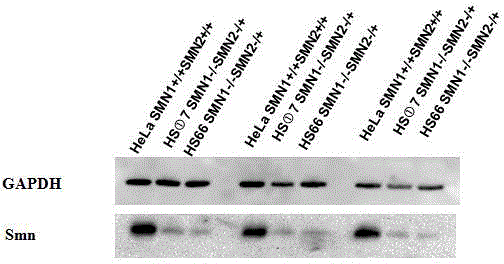
[0097] For the obtained two SMN1- / - SMN2+ / - genotype Hela cell lines HS-66 and HS-1-7, when they grow to 80-90% in a 60mm cell culture dish, trypsinize, lyse the cells, and extract Protein, use a microplate reader to measure the protein concentration, take 45ug of the sample and load it on SDS-PAGE electrophoresis, use 10% separating gel, 5% stacking gel, PVDF membrane wet transfer, transfer time 1h, rabbit anti-SMN Antibody monoclonal antibody incubation 1h, Goat -Anti-rabbit secondary antibody was incubated for 1 hour, detected by ECL fluorescence, developed and fixed, and photographed. as attached Figure 4 As shown, compared with normal HeLa cells, the amount of SMN protein expressed by the SMN gene knockout cells was significantly reduced.
[0098]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com