3-methylthio tyrosine translation system and application thereof
A technology of methionine and amino acid, which is applied in the field of biochemistry and can solve problems such as inability to detect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
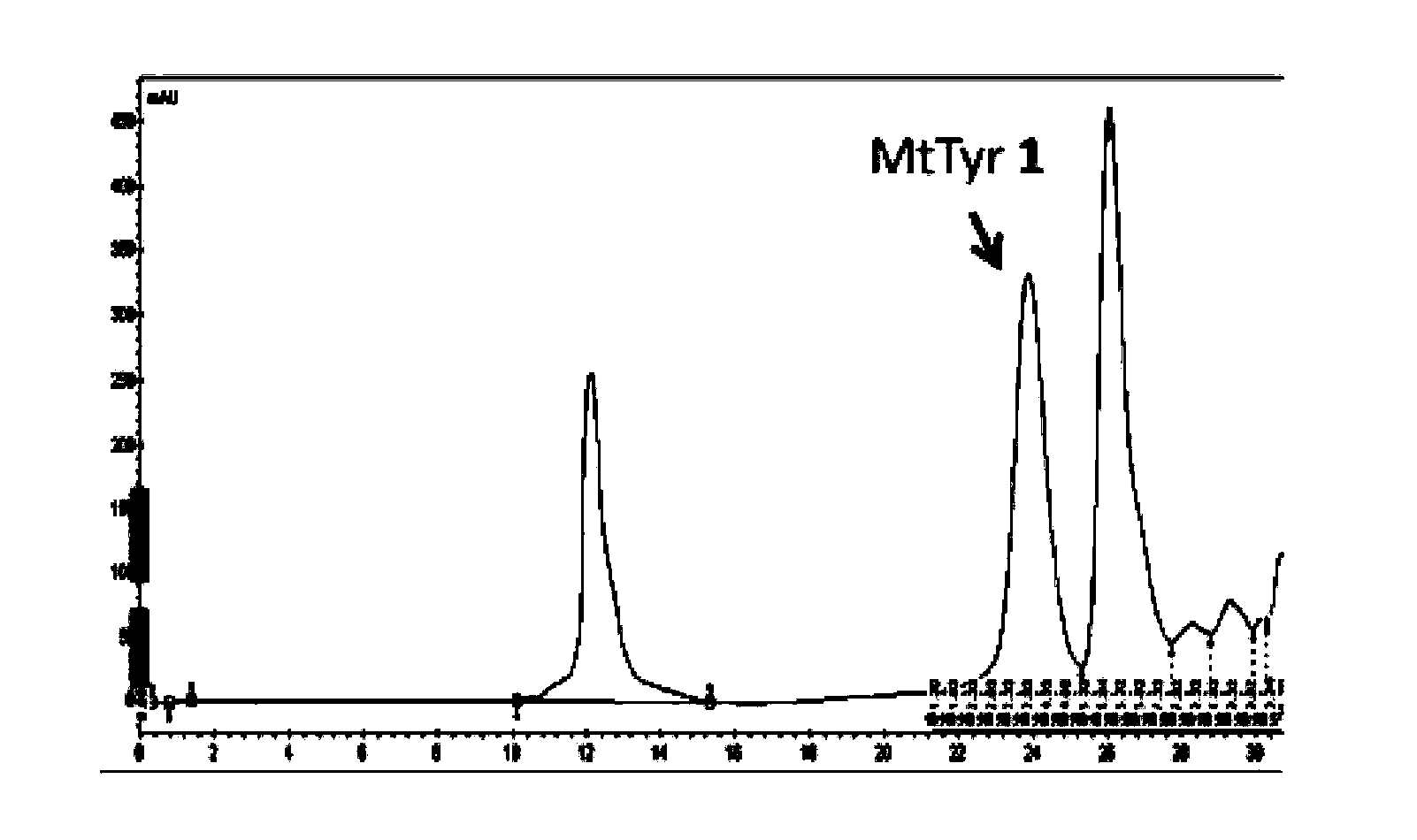
[0077] Example 1: Biocatalytic synthesis of MtTyr ( Figure 1-Figure 4 )
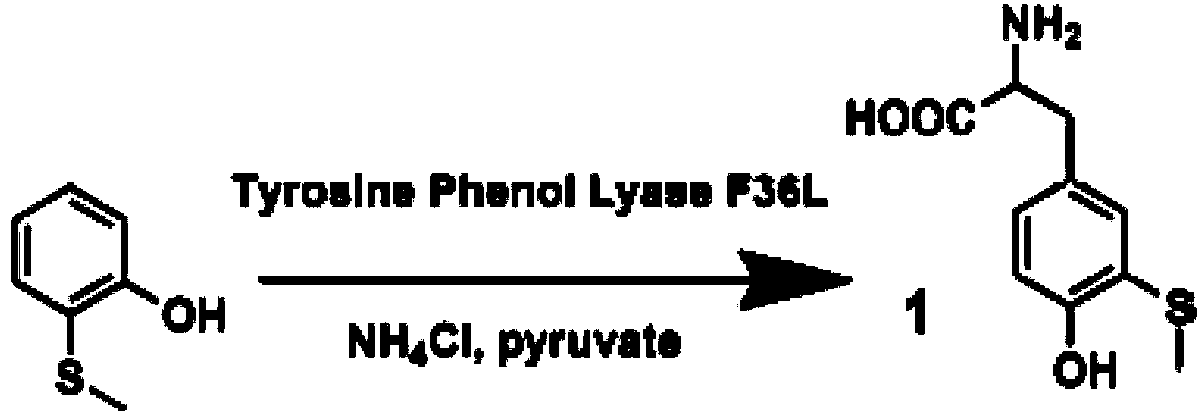
[0078] The present invention adopts the method of biological enzyme catalysis, using tyrosine phenol lyase (TPL) cloned from Citrobacter Freundii (ATCC 8090, purchased from the American Type Culture Collection (ATCC)) to catalyze 2-hydroxybenzene Synthesis of 3-Methylthiotyrosine from thioether, see the catalytic reaction formula figure 1 .
[0079] However, experimental results show that wild-type TPL cannot catalyze the production of 3-methylthiotyrosine. Therefore, the inventors analyzed the crystal structure diagram of TPL ( figure 2 A), select 448-position phenylalanine, 36-position phenylalanine and 288-position methionine, introduce NNK mutation (N=A+T+C+G; K=T+G), construct pEt -TPL mutation library for directed evolution of TPL. 96 single clones were selected from the mutant library and cultured overnight in a 96-well plate. Lysozyme was added to lyse the cells, and then 2-hydroxyphenyl sulfide, ...
Embodiment 2
[0083] Example 2: Evolution of MtTyr specific aminoacyl-tRNA synthetase
[0084] In order to specifically insert MtTyr into the gene, it is necessary to introduce the aminoacyl-tRNA synthetase / tRNA orthogonal pair into the E.coli host cell used. This orthogonal pair is derived from Methanococcus jannaschii amber inhibition Tyrosyl tRNA (MjtRNA CUA Tyr ) / Tyrosyl tRNA synthetase (MjTyrRS, wild type, whose amino acid sequence is SEQ ID NO: 2) pair. The MjTyrRS mutation library was constructed in a kanamycin-resistant pBK plasmid (purchased from the Peter G. Schultz laboratory of the Scripps Research Institute, USA), located between the promoter and terminator of E. coli glutamine synthetase on the plasmid. The synthetase mutation library used is the pBk-lib-jw1 library, and the method for constructing the mutation library is to select 6 sites (Tyr32, Leu65, Phe108, Gln109, Asp158, and Leu162) on the MjTyrRS gene to introduce NNK mutations N=A+T+C+G; K=T+G), the other 6 sites (Ile...
Embodiment 3
[0088] Example 3: Expression of MtTyr-myoglobin and identification by mass spectrometry
[0089] The orthogonal tRNA (SEQ ID NO: 1) and the nucleotide sequence (4TAG or 33TAG) encoding myoglobin (SEQ ID NO: 5 and 7) were constructed into the pBAD vector (purchased from the American Scripps Research Institute Peter G. Schultz) Laboratory), the selected nucleotide sequence encoding MtTyrRS (SEQ ID NO: 3) was constructed on pBK vector (purchased from the Peter G. Schultz laboratory of the Scripps Research Institute, USA), and then co-transformed into DH10B cells (purchased From the full gold company). Pick a single clone and culture it to OD at 37°C 600 When approximately equal to 0.5, 1mMMtTyr and 0.2% arabinose (purchased from sigma company) were added to the LB medium to culture the cells, and the control did not add MtTyr. After 6-8 hours, the bacteria were collected, the protein was purified by Ni-NTA, and analyzed by SDS-PAGE electrophoresis ( Image 6 A).
[0090] We found th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com