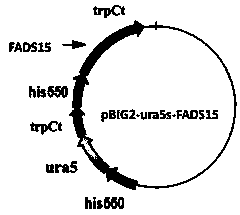
A Mortierella alpina genetic engineering strain overexpressing ω3 desaturase gene and its construction method
A genetically engineered strain, Mortierella alpina technology, applied in the field of bioengineering, can solve the problems of insufficient activity, low EPA production, and low expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Cloning of the Mortierella alpina ω3 desaturase gene (FADS15)
[0040] According to the sequence information of the Mortierella alpina ω3 desaturase gene (FADS15), primers P1 and P2 were designed, and the underlined parts are the restriction sites NheI and SacI, respectively, for the plasmid pET19b-FADS15 containing the ω3 desaturase gene (FADS15) as a template (this lab has published articles Haiqin Chen, Zhennan Gu, Hao Zhang, Mingxuan Wang, WeiChen, W.Todd Lowther, Yong Q.Chen*.Expression and Purification of Integral Membrane Fatty Acid Desaturases.PLoS ONE.2013,8( 3): e58139), using primers P1 / P2 and KOD high-fidelity polymerase to amplify the ω3 desaturase gene (FADS15) by PCR to obtain a fragment. The PCR program was: 94°C for 30s, 55°C for 30s, 68°C for 1.5min, 30 cycles, and the PCR product was purified, and the purified product was verified by 1.2% agarose gel electrophoresis.
[0041] P1 (sense): GCCATA GCTAGC AAATGGCACCCCCTCACGTTGT
[0042] P2 ...
Embodiment 2
[0044] Embodiment 2: Construction of binary expression vector
[0045] 1. Enzyme digestion reaction
[0046] Under the condition of 37°C, the restriction endonuclease NheI was first digested overnight, the product ω3 desaturase gene fragment FADS15 and the vector pBIG2-ura5s-ITs fragment were purified by PCR, and the gel was recovered. The NheI digestion system (100 μL) is: 2 μL NheI-HF, 1 μL BSA, 30 μL plasmid or PCR product, 10 μL Buffer4 (purchased from NEB Company, Cat. No. B7004S), 57 μL deionized water, digested in a water bath at 37°C for 5 hours.
[0047] The digested product was recovered and digested with SacI. The enzyme digestion system was (100 μL): 2 μL SacI, 30 μL plasmid or PCR product, 10 μL Buffer4 (purchased from NEB Company, Cat. No. B7004S), 58 μL deionized water, digested in a 37°C water bath 5h.
[0048] 2. Ligation reaction
[0049] The digested ω3 desaturase gene fragment FADS15 was ligated with the vector pBIG2-ura5s-ITs with T4 ligase, and ligated...
Embodiment 3
[0058] Example 3: Agrobacterium tumefaciens-mediated transformation of MAU1 (CCFM501)
[0059] On the basis of the existing domestic and foreign literature reports on the transformation method of Agrobacterium tumefaciens, appropriate optimization and adjustments have been made. The specific successful implementation examples are as follows:
[0060] (1) Agrobacterium tumefaciens C58C1 containing the plasmid pBIG2-ura5s-FADS15 stored at -80°C was streaked on a YEP solid medium plate containing 100 μg / mL rifampicin and 100 μg / mL kanamycin. Incubate in the dark at 30°C for 48 hours.
[0061] (2) Pick a single clone and inoculate it into 20 mL of MM liquid medium containing 100 μg / mL rifampicin and 100 μg / mL kanamycin at 30°C, 200 rpm, and incubate in the dark for 24-48 hours.
[0062] (3) Centrifuge at 4000g for 5 minutes to collect the bacteria, and discard the supernatant. Add 5mL IM liquid medium to resuspend the bacteria, centrifuge at 4000g for 5 minutes, and discard the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com