Expression equipment and genetically engineered bacterium for expressing foreign proteins in penicillium expansum cells
A technology of genetically engineered bacteria and Penicillium expanded, applied in bacteria, biochemical equipment and methods, fungi, etc., can solve the lack of post-translational processing and folding mechanism, the difficulty of active proteins, and the complex renaturation of inclusion bodies, etc. problems, to achieve the effect of easy preservation and DNA manipulation, high matching degree and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
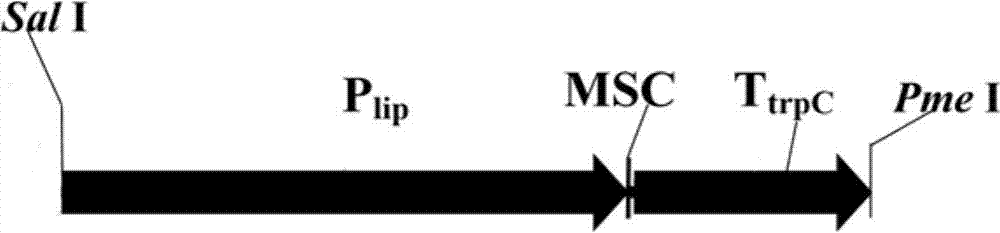
[0061] Example 1, Preparation of Extended Penicillium Heterologous Expression Equipment
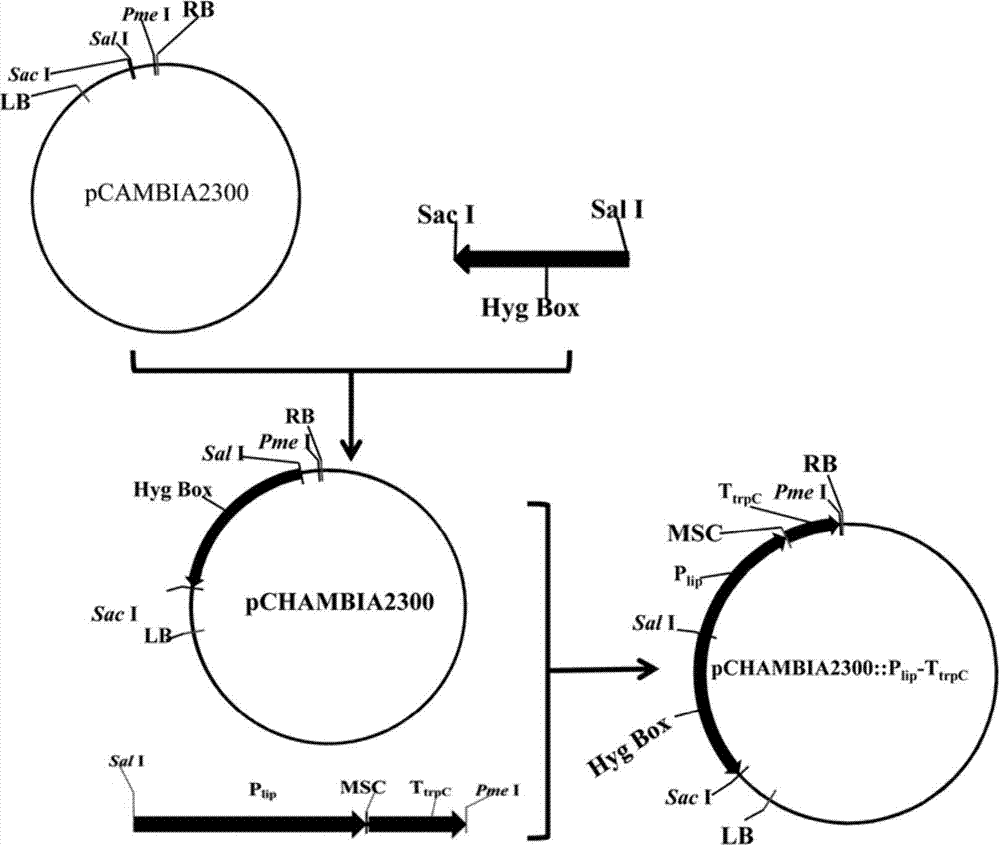
[0062] (1) Utilize restriction endonuclease Sac 1, Sal I to convert hygromycin resistance expression cassette from plasmid pV2 (Wang Y, Guo B, Miao Z, Tang K. Transformation of taxol-producing endophytic fungi by restriction enzyme-mediated integration (REMI). FEMS Microbiology letter, 2007, 253-259), the fragment was recovered by gel, and cloned into the corresponding restriction site of pCAMBIA2300 plasmid to obtain the recombinant plasmid pCHAMBIA2300 with hygromycin resistance; The hygromycin expression cassette can also be derived from any other DNA containing the sequence, wherein the nucleotide sequence of the hygromycin phosphotransferase gene is shown in SEQ ID NO:10.
[0063] (2) Take 0.2g of Penicillium expansica mycelium, grind it with liquid nitrogen, add 1mL of CTAB extract, bathe in water at 65°C for 30min, centrifuge at 12000rpm / min for 10min, take the supernatant; add 0...
Embodiment 2
[0078] Embodiment 2, the acquisition of expanding Penicillium genetically engineered bacteria
[0079] The detailed steps are as follows:
[0080] (1) Pick isolated and purified wild-type Penicillium expansica and inoculate it on a PDA plate, culture it at 28°C for about 20 days, and wash the mature spores with sterile water.
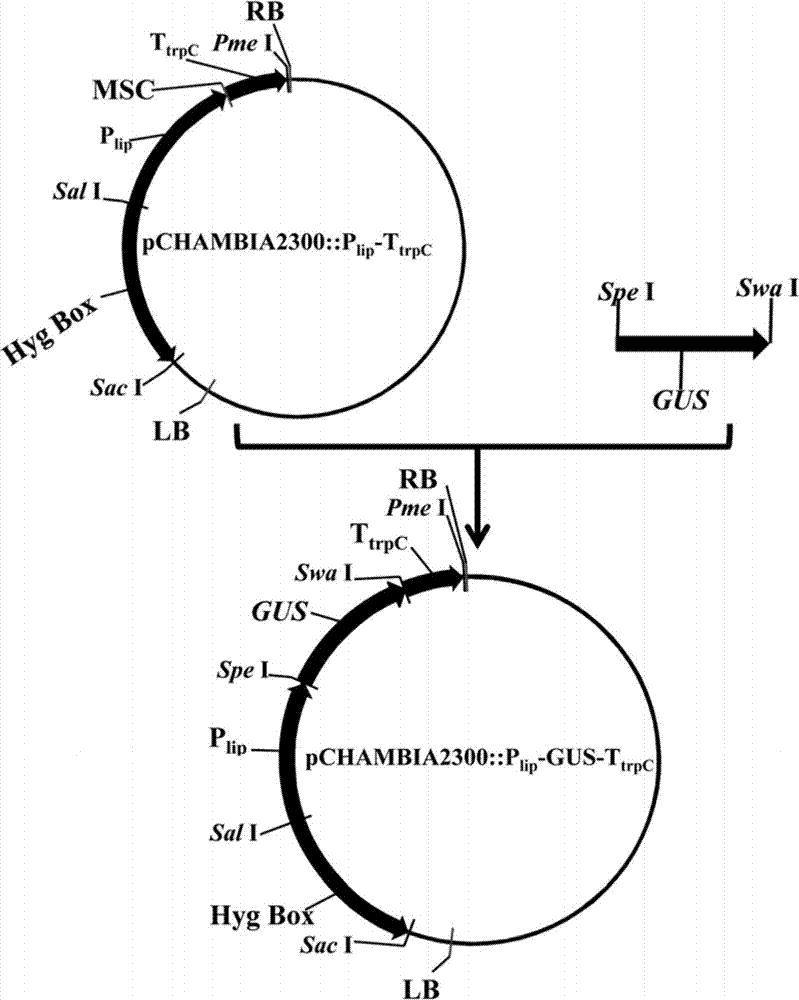
[0081] (2) the final vector pCHAMBIA2300 containing in Example 1:: P lip -GUS-T trpC The engineered Agrobacterium EHA105 was inoculated in LB liquid medium containing 100 μg / mL streptomycin and 100 μg / mL kanamycin at 28°C and 200 rpm for overnight culture, and was cultured overnight with 100 μg / mL streptomycin and 100 μg / mL kanamycin The MM culture medium was reactivated, and cultured at 28°C and 220rpm for 48 hours. Draw an appropriate amount of culture and centrifuge at 5000rpm to remove the supernatant, wash with 1M liquid medium, and finally dilute to OD with 1M liquid medium 600 =0.15, then cultured at 28°C, 220rpm for 6-8 hours, until OD 60...
Embodiment 3
[0083] Example 3, Production of GUS protein using Penicillium expanses expression equipment
[0084] Inoculate the genetically engineered Penicillium extensa and the wild-type Penicillium expanse obtained in Example 2 into GM medium respectively, shake and culture at 220 rpm and 26° C. for 2 days, and then filter and recover mycelia. The mycelium was washed three times with distilled water, then put into an eppendorf tube, added GUS dye solution, incubated at 37°C for 3-5h, and the staining of the mycelium was observed. See Figure 4 , Figure 4 Shows transferred into pCHAMBIA2300::P lip -GUS-T trpC The genetic engineering strain of Penicillium expanses presents a remarkable blue color ( Figure 4 II, III, IV in ), while the wild-type Penicillium expanses strain was not stained ( Figure 4 in I). The results show that after using the expression equipment of the present invention, the exogenous protein gene GUS can be highly expressed in the Penicillium expanses cells. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com