Primer and probe for alicyclobacillus acidoterrestris PCR detection and applications thereof
A technology of Bacillus and alicyclic acid, which is applied in the field of food microbial detection, can solve the problems of unapplied, low content, and detection lag, and achieve broad application prospects, high sensitivity, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Primer and probe design and synthesis
[0020] Use Primer Premier5 software to compare the 16SrDNA gene sequences of 10 different species in Alicyclobacillus spp., such as figure 1 As indicated, primers and fluorescent probes based on the conserved region of A. acidoterrestris were designed using the probe design software Primer Express, as shown in Table 1. After extensive screening, the primer sequences and probes of the following sequences were obtained, as shown in Table 2.
[0021] The upstream primer (FP) sequence of the primer is SEQ ID NO.1, and the downstream primer (RP) sequence of the primer is SEQ ID NO.2. The composition of the specific primer sequence is as follows:
[0022] SEQ ID NO.1: 5'-TGAGT AACAC GTGGG CAATC TG-3';
[0023] SEQ ID NO. 2: 5'-CTACC CGTGT ATTAT CCGGC AT-3'.
[0024] The nucleotide sequence of the fluorescent probe is: 5'-CTTTCAGACTGGAATAAC-3', the 5' end of the probe is labeled with 6-carboxyfluorescein (FAM) fluorescent dye, and the...
Embodiment 2
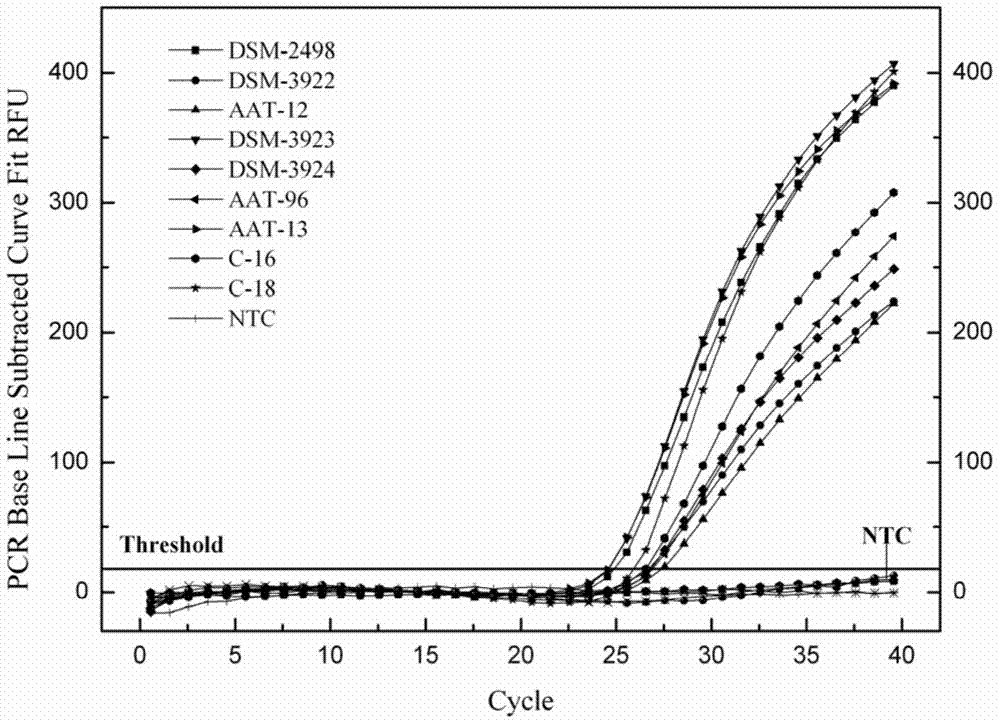
[0035] specificity test
[0036] The real-time fluorescent PCR detection system that adopts primer SEQ ID NO.1, SEQ ID NO.2 and probe SEQ ID NO.3 to set up in the method of the present invention detects bacterial strain as shown in table 1, evaluates primer, probe among the present invention The specificity of needle and real-time fluorescent PCR detection system.
[0037] The 30 bacterial strains shown in Table 1 were cultured under optimal conditions respectively, and their concentration was adjusted to 10 with sterile ultrapure water. 4 -10 5CFU / mL. Place 5 mL of the prepared bacterial cell dilution sample in a centrifuge tube, freeze and centrifuge at 4°C and 6000 rpm for 10 min, and discard the supernatant. Add 20mg / mL of lysozyme buffer (20mmol / L Tris-HCl, pH8.0, 2mmol / L EDTA and1.2%Triton X-100) and treat in a water bath at 37°C for 30min. Extract according to the instructions of the kit, and finally dissolve the extracted sample with 100 μL eluent. Adopt nucleic a...
Embodiment 3
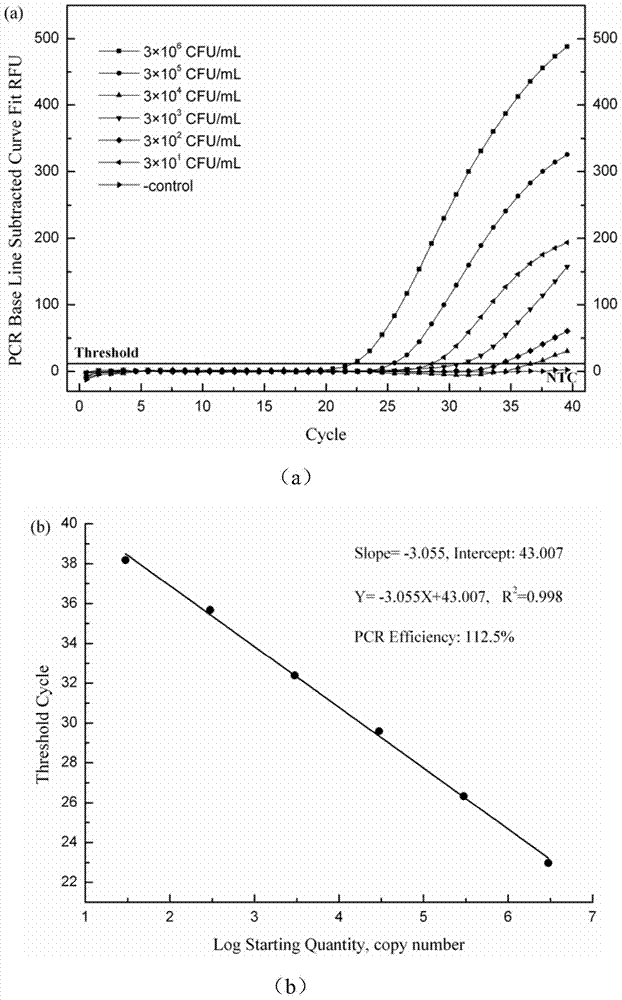
[0041] Sensitivity experiment
[0042] Take 10mL A.acidoterrestris standard strain proliferation medium (3 × 10 7 CFU / mL), diluted 10 times with sterile ultrapure water, the concentrations were: 3×10 6 CFU / mL, 3×10 5 CFU / mL, 3×10 4 CFU / mL, 3×10 3 CFU / mL, 3×10 2 CFU / mL, 3×10 1 CFU / mL, 3×10 0 CFU / mL. Place 5 mL of the prepared bacterial cell dilution sample in a centrifuge tube, freeze and centrifuge at 4°C and 6000 rpm for 10 min, and discard the supernatant. Add 20mg / mL of lysozyme buffer (20mmol / L Tris-HCl, pH8.0, 2mmol / L EDTA and1.2%Triton X-100) and treat in a water bath at 37°C for 30min. Extract according to the instructions of the kit, and finally dissolve the extracted sample with 100 μL eluent. A nucleic acid titrator is used to measure the concentration and purity of the extracted DNA, and the PCR amplification detection of the sample is carried out according to the above-mentioned method. At the same time, according to the relationship between the sample co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com