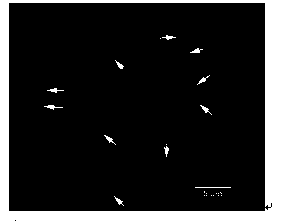
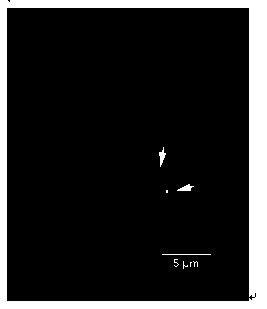
Fluorescence in situ hybridization method of 5S rDNA on plant chromosome
A fluorescence in situ hybridization and chromosome technology, applied in the field of cytogenetics and molecular biology, to achieve the effect of simple method, simplified pretreatment steps, and simple elution steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] FISH Mapping of 5S rDNA on the Metaphase Chromosome
[0052] 1. Chromosomal Specimen Preparation
[0053] (1) The root tip of mustard mustard seeds grows to 0.5cm after germination.
[0054] (2) Cut off the root tip and pretreat it with saturated p-dichlorobenzene aqueous solution for 5 hours at room temperature.
[0055] (3) The root tip was hypotonic with 0.075mol / L potassium chloride hypotonic solution for 30 minutes, and then fixed with 3:1 (methanol: glacial acetic acid) for 20 minutes.
[0056] (4) Wash three times with distilled water, 5min each time, to fully wash away the fixative.
[0057] (5) Using 2.5% (w / w) pectinase and cellulase mixed in a weight ratio of 1:1, the root tip was dissociated at 37°C for 1 hour.
[0058] (6) Wash off the enzyme solution with distilled water, hypotonic with 0.075mol / L potassium chloride hypotonic solution for 30min, and fix with 3:1 methanol: glacial acetic acid fixative for 20min.
[0059] (7) Prepare chromosome specimens...
Embodiment 2
[0082] Embodiment 2: comparative embodiment
[0083] classic in situ hybridization
[0084] step:
[0085] Steps for FISH mapping of 5S rDNA on the metaphase chromosomes of potherb mustard:
[0086] 1. Chromosomal Specimen Preparation
[0087] (1) The root tip of mustard mustard seeds grows to 0.5cm after germination.
[0088] (2) Cut off the root tip and pretreat it with saturated p-dichlorobenzene aqueous solution for 5 hours at room temperature.
[0089] (3) The root tip was hypotonic with 0.075mol / L potassium chloride hypotonic solution for 30 minutes, and then fixed with 3:1 (methanol: glacial acetic acid) for 40 minutes.
[0090] (4) Wash three times with distilled water, 5min each time, to fully wash away the fixative.
[0091] (5) Using 2.5% (w / w) pectinase and cellulase mixed in a weight ratio of 1:1, the root tip was dissociated at 37°C for 1 hour.
[0092] (6) Wash off the enzyme solution with distilled water, hypotonic with 0.075mol / L potassium chloride hyp...
Embodiment 3
[0125] Examples of practical applications
[0126] Application method of the present invention has been in such as poplar (see image 3 ), cauliflower (see Figure 4 ),carrot( Figure 5 ) and more than 30 kinds of plants have been successfully tested on a variety of plants,
[0127] Method: with embodiment 1
[0128] Step: with embodiment 1
[0129]Conclusion: The method of the present invention can be extended and applied to 5S rDNA FISH localization of various plants.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com